Working with large and complex molecular systems can often feel overwhelming, especially when you’re trying to isolate a specific part of your model—ligands, active sites, or particular side chains—for editing, analysis, or visualization. If you’ve ever found yourself switching endlessly through chains and structures to pinpoint just the atoms or residues you need, you’re not alone.
To help tackle this, the Node Specification Language (NSL) in SAMSON offers a feature that can significantly streamline your workflow: filtering nodes using the Document View. This lets you write simple (and also expressive) queries to display only the elements that match your current goal.
What is the Document View?
The Document View in SAMSON displays all elements—molecules, atoms, structures—of your current model. It’s like your table of contents for the entire system. Normally, it’s easy to navigate but gets harder as the model grows. Filtering helps reduce visual overload so that you can focus only on the relevant subset.
Using NSL Expressions in Document View
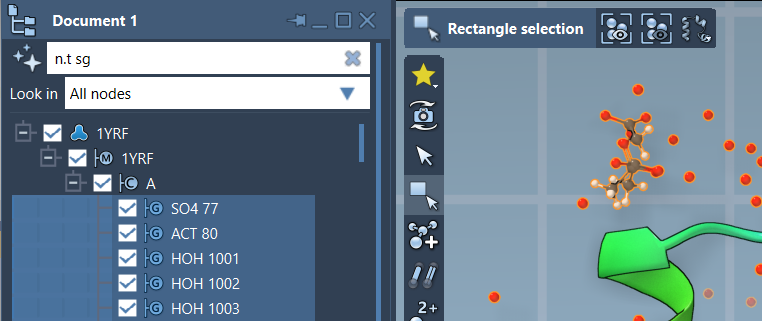
To filter nodes in the Document View, simply type an NSL expression into the filter field and press Enter. For example, if you want to see only structural groups, you can type:
|
1 |
n.t sg |
This is shorthand for:
|
1 |
node.type structuralGroup |
Once you’ve filtered what’s visible, you can also select these nodes directly by clicking Enter again. This makes it easier to perform operations like hiding, modifying, or exporting just those nodes.
Why Is This Useful?
- 🔍 Quick selection: Instead of manually checking boxes or digging through hierarchies, type what you need.
- 🧠 AI assistance: Stuck? Ask the AI Assistant in SAMSON to generate NSL expressions based on your document.
- 💡 Flexible expressions: Combine with logic and topology—for example only selecting backbones in residues with certain IDs.
Example queries you might find useful:
r.id 30:40: Show residues between IDs 30 and 40a.x -1nm:1nm: Display atoms with X-coordinates in [-1nm, +1nm]n.t sc h S: Show side chains containing sulfur atoms
This targeted approach saves time and cognitive effort, particularly when working on detailed structural analysis or complex biomolecular assemblies.
Learn More
To dive deeper into the full capabilities of NSL expressions, including advanced logical filters and proximity operations, check the full Node Specification Language documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. Get SAMSON here.





