When designing new molecules, especially in early-stage drug discovery, chemists often need to generate analogs by modifying specific parts of a lead compound. This process, known as isosteric replacement or fragment replacement, can be extremely tedious and laborious. Doing this manually or with scripts can be error-prone and time-consuming, especially when managing complex molecules and keeping track of dozens – even hundreds – of variants.
If you’ve ever wondered whether there’s a better way to manage this process visually and interactively, the SMILES Manager in SAMSON might be just what you’re looking for. One of its powerful but sometimes overlooked features is the Replace fragments tab, which streamlines the generation of focused molecular libraries based on SMARTS-defined fragment substitution. And it’s all done within an intuitive interface.
How does it work?
Let’s imagine you have a known active molecule and you want to explore how changing a specific chemical group affects its properties. Here’s how you can quickly do that in the SMILES Manager:
- Input your initial molecule: You can load it from file, paste it, or import it directly from your current SAMSON document. For example:
COc1cc(CNC(=O)CCCC/C=C/C(C)C)ccc1O - Specify the fragment to replace: Use SMARTS notation to select the substructure to substitute. A typical example might be:
[*:1]C(=O)NC[*:2] - Add replacement fragments: These should also follow the SMARTS pattern syntax with two wildcards. For example:
[*:1]S(=O)(=O)NC[*:2],[*:1]C1=NN=C([*:2])N1, and many others. - Click ‘Replace fragments’ and watch SAMSON automatically generate a full library of resulting molecules, populating a structured table with names and 2D depictions.
Each generated compound can then be exported to your SAMSON document, translated into a 3D structure, or saved as individual or grid images for further analysis.
Why this matters
This workflow is especially helpful when you’re trying to quickly:
- Explore bioisosteric replacements
- Design analogue series for SAR studies
- Create custom fragment libraries for virtual screening
- Visualize how changes impact the overall molecule before jumping into computational simulations
Because the replacement process is reactive—updating names and images as you go—you can catch errors early and keep clean, well-labeled data.
One file becomes dozens of candidate molecules
All it takes is a few clicks and your single starting compound can become an entire family of analogs. Paired with 3D structure generation and customizable grid output, this makes the Replace Fragments feature much more than a tool—it’s a workbench for idea generation.
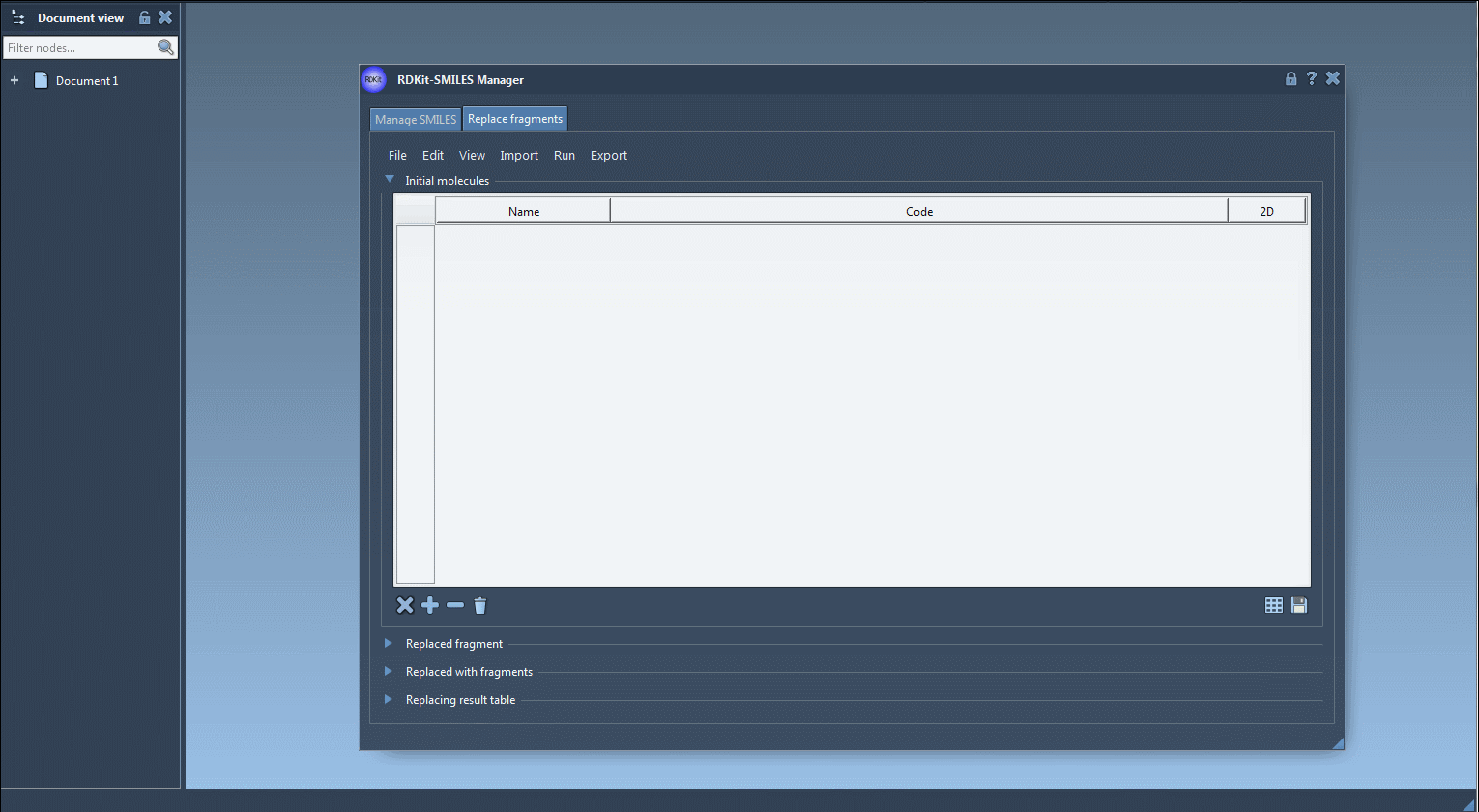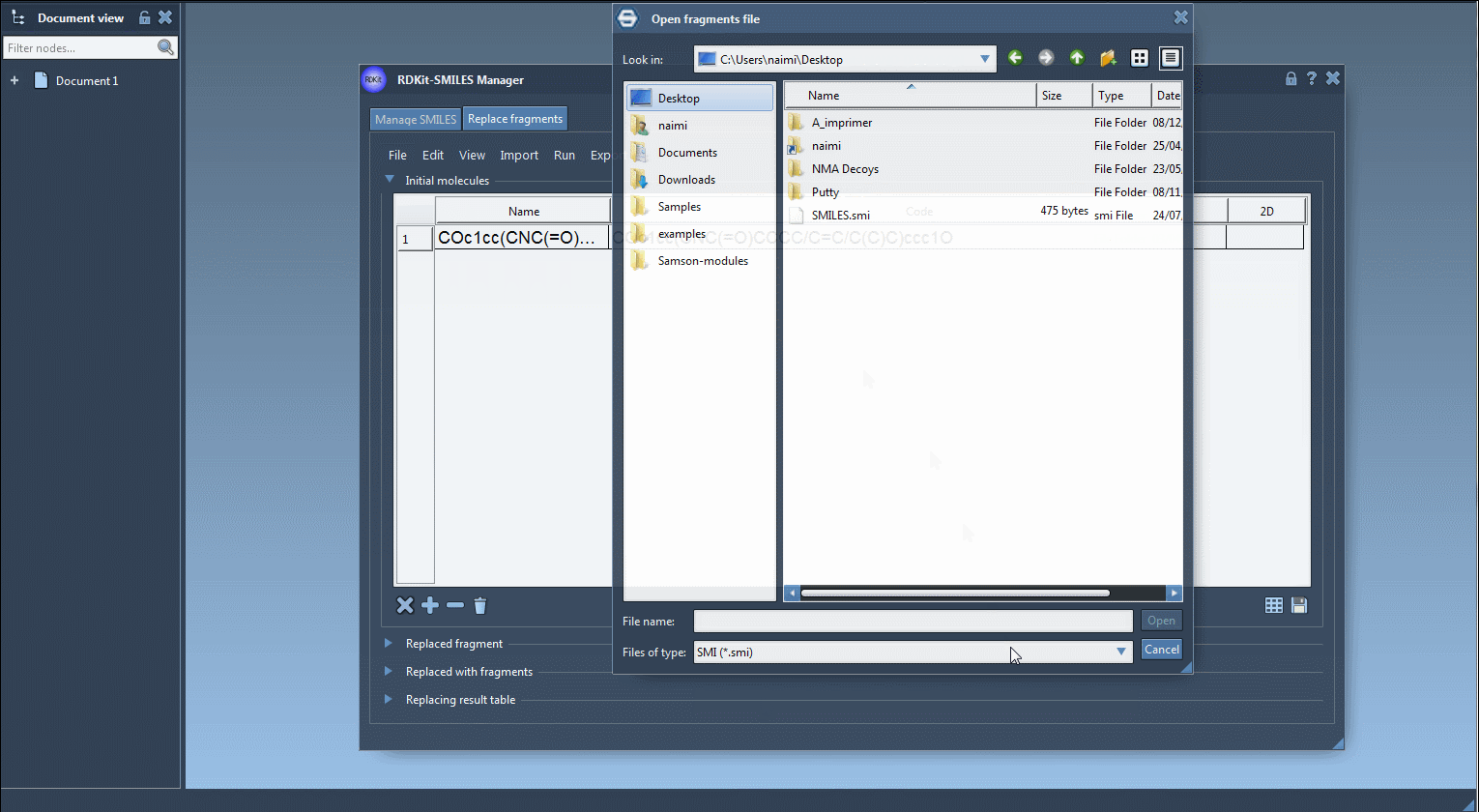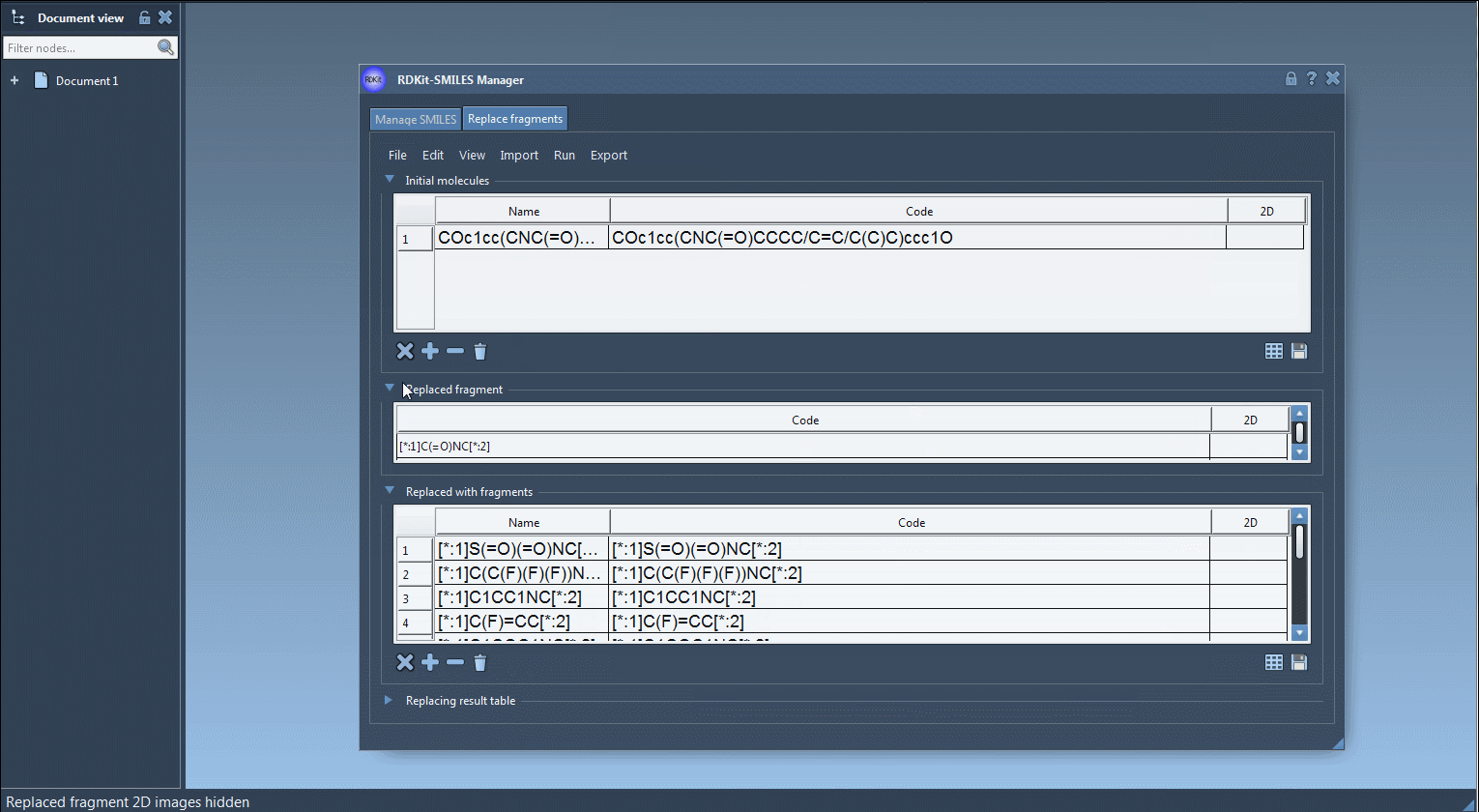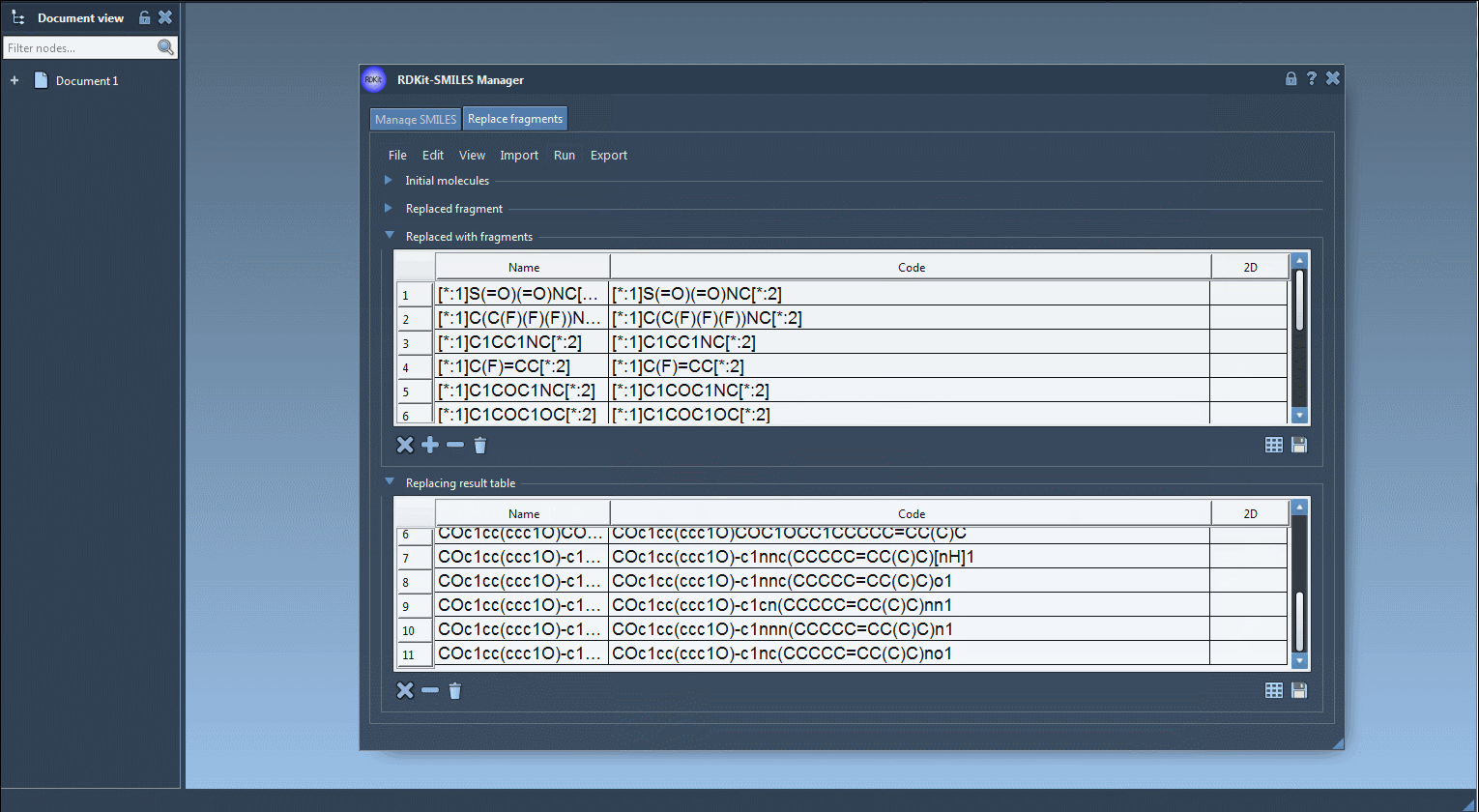
You can then view the results, examine the automatically generated depictions, and decide which to pursue further or export for more detailed modeling.
To get started using this feature, you can follow the in-depth steps provided in the official documentation. It’s clear and well-illustrated, whether you’re experimenting with a few fragments or building an entire combinatorial library.
👉 Learn more in the official documentation
Note: SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at www.samson-connect.net.





