For molecular modelers working on viral proteins, computing conformational transitions between known structures is a frequent challenge. Whether designing potential inhibitors, studying viral entry, or simply trying to understand structural dynamics, one practical problem remains: how to interpolate between two experimentally known states when their residue counts differ?
This scenario appears when working on SARS-CoV-2’s infamous spike protein. The spike shifts from a closed (inactive) conformation to an open (active) conformation to bind to human cells through ACE2 receptor docking. Structures for both states exist (PDB 6VXX and 6VYB), but they differ slightly, creating a computational challenge for generating smooth conformational transitions.
Animating an asymmetric bio-machine
Using SAMSON, researchers can calculate a plausible transition between closed and open states using structure-based interpolation. The spike protein is not a simple rigid body — its deformations are highly asymmetric and nuanced. The steps below detail how scientists at OneAngstrom bridged this in silico using two powerful SAMSON modules: ARAP and P-NEB.
Steps summary:
- Preprocessing: Sugar bond orders were corrected via Python for compatibility with hydrogen addition and minimization.
- Minimization: Both open and closed state models were fully hydrogenated and energy-minimized to ensure stable baselines.
- ARAP Interpolation: A smooth mathematical path was generated between open and closed states with the ARAP Interpolation Path module—ready in under 30 seconds on a laptop.
- P-NEB Refinement: With small structural residue differences in mind, a closed-state structure was extracted from the ARAP path and used as a new goal state. This was then reprocessed through the P-NEB module to yield a refined and more biochemically plausible transition.
What does it look like?
The result: a vivid and revealing animation of the spike protein transitioning between its two primary conformations—a powerful visual tool for both research and education.
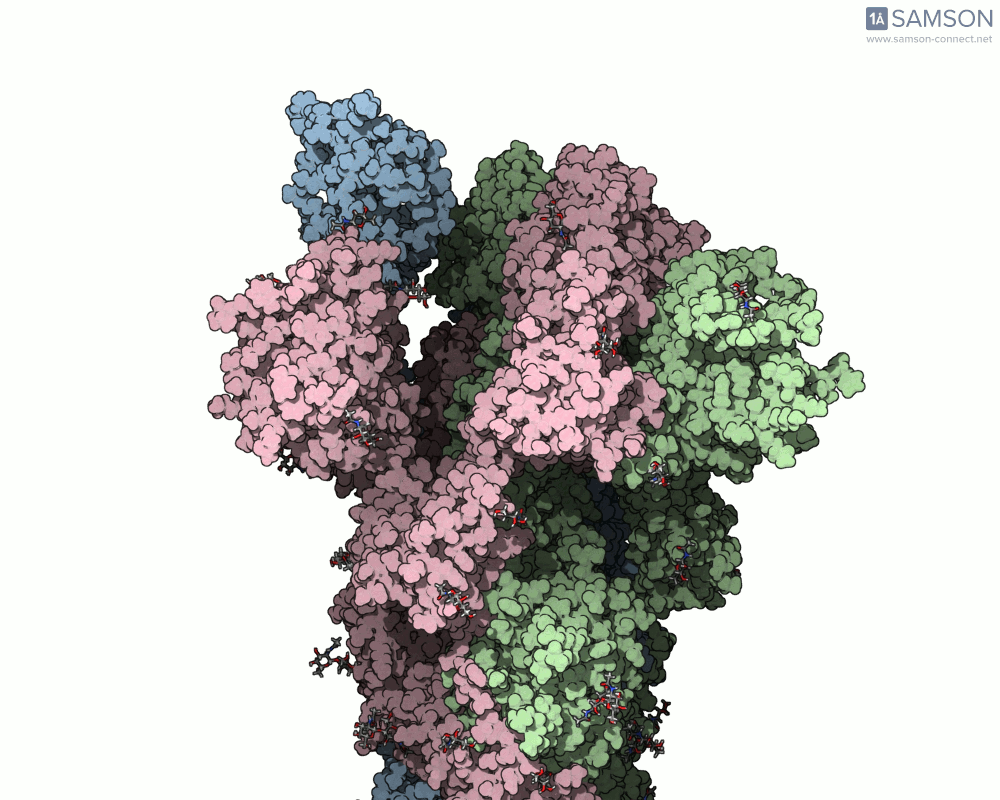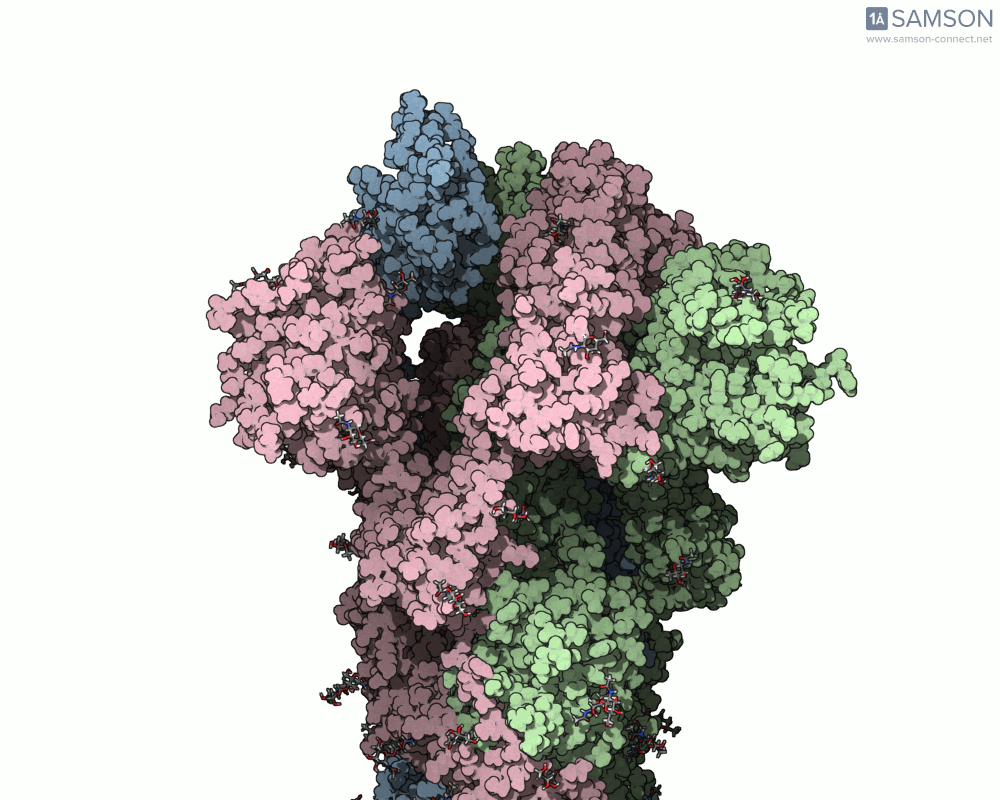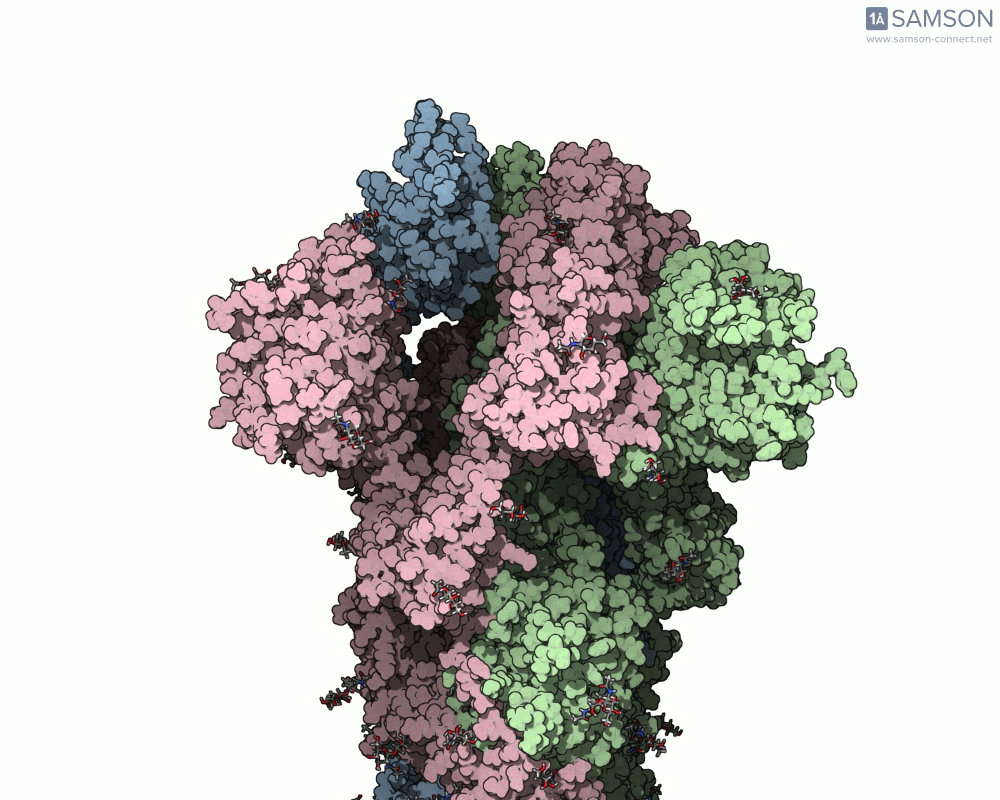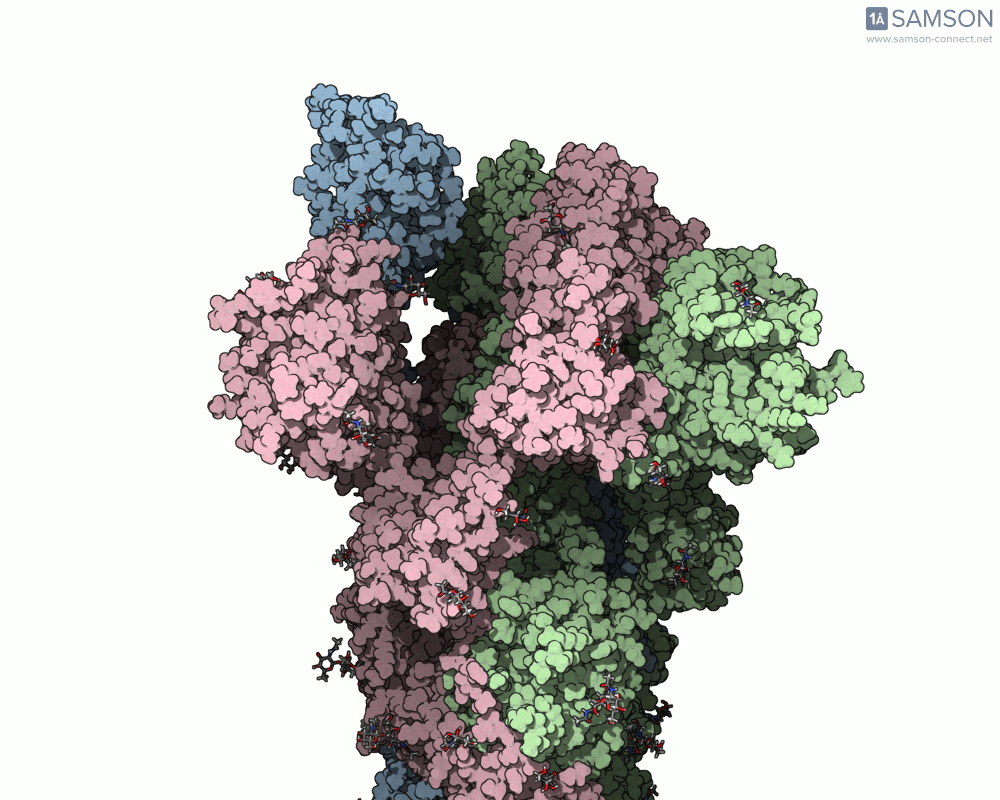
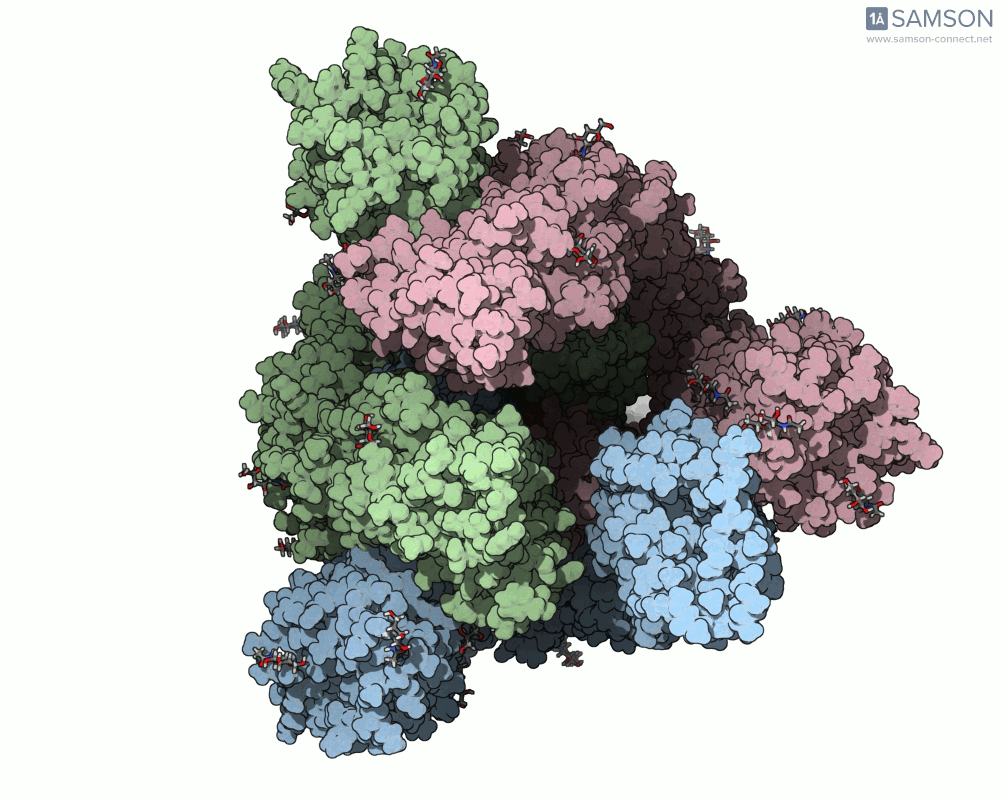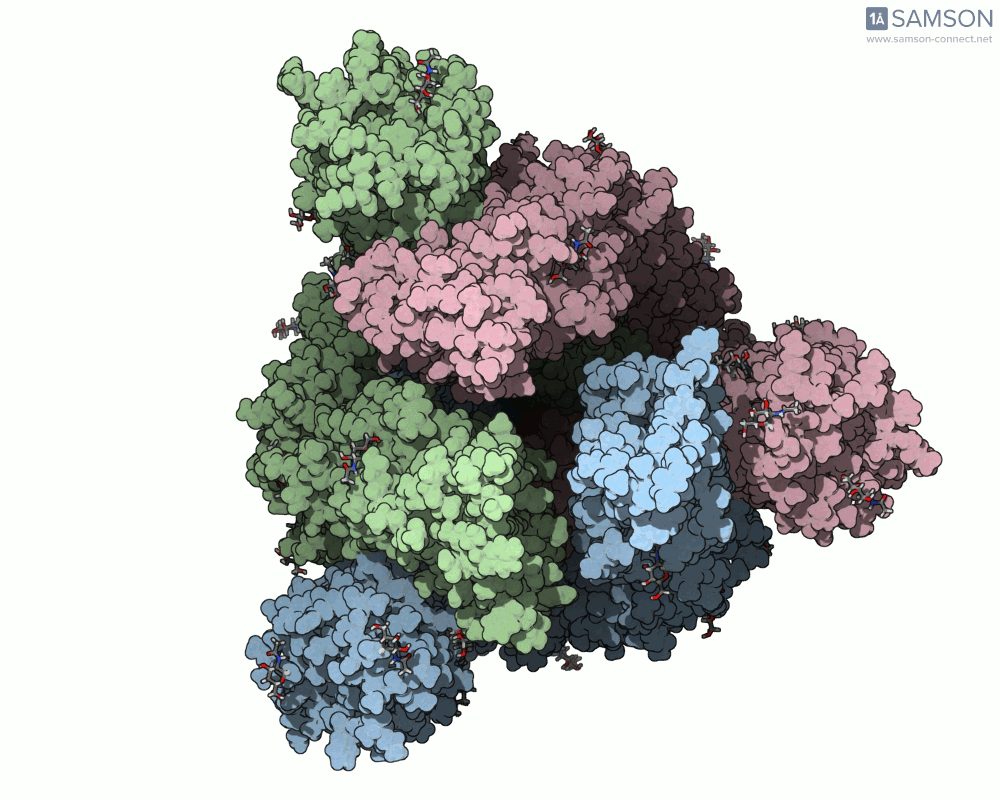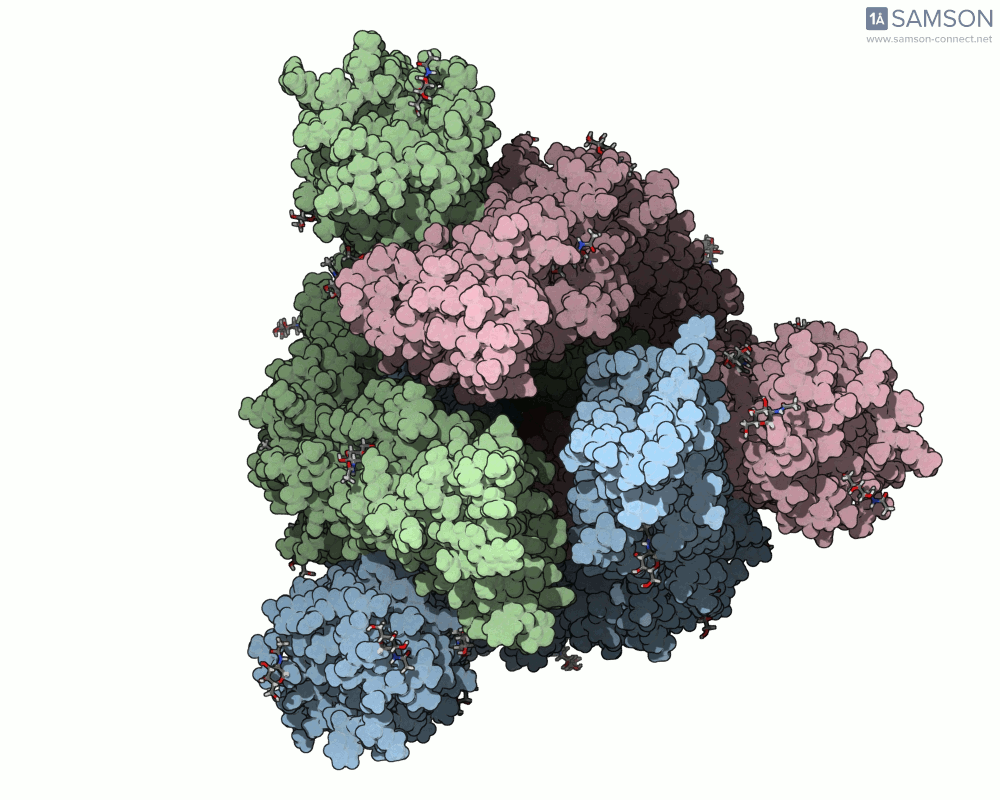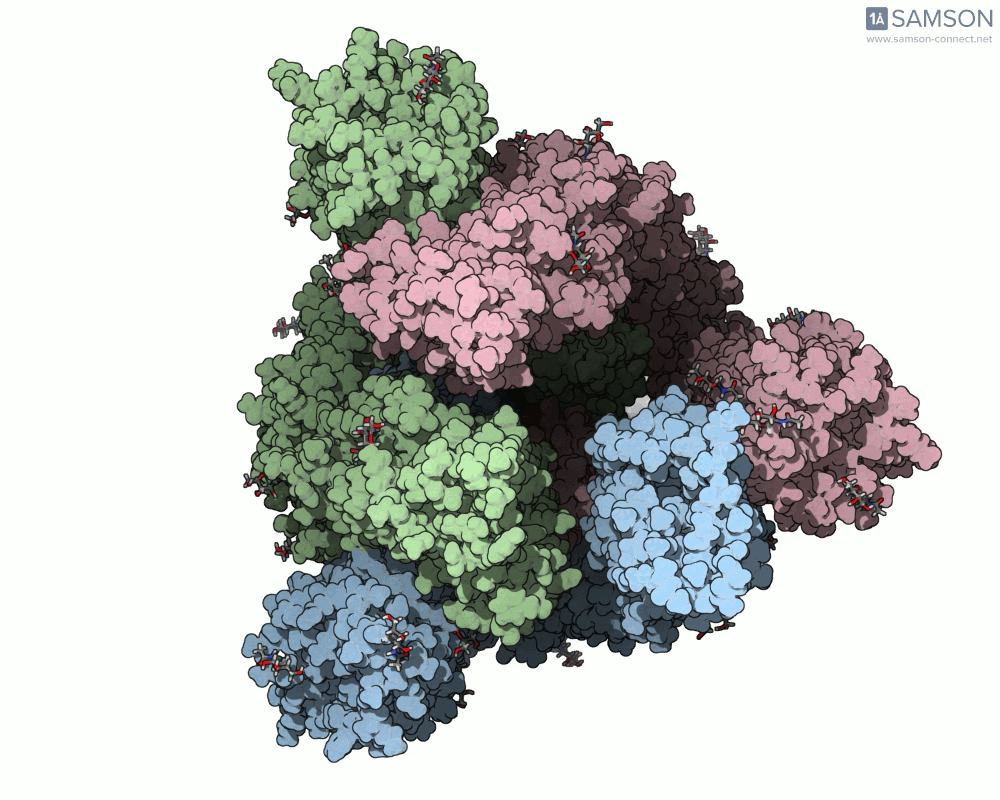
Downloads available
The computed conformational pathway is available in several formats:
These facilitate structural analysis, RMSD comparisons, or custom visualization in other molecular modeling environments.
Why it matters
Interpolating transitions between biological macrostates is critical when studying conformational energy landscapes or training machine learning models on biological motion. If data consistency is compromised by missing residues or incompatible files, SAMSON’s ARAP and P-NEB pipeline offers a relief: a reproducible method, even tolerant to structural inconsistencies, that swiftly delivers interpretable and shareable animations.
To dive deeper into how this specific conformational transition was computed and learn more about structural biology workflows in SAMSON, consult the full tutorial: SARS-CoV-2 Spike Motion Documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





