Reaction coordinate studies and free energy calculations can be challenging, especially when it comes to managing atomic trajectories. For molecular modelers, exporting precise atomic coordinates along defined paths is a crucial but potentially time-intensive task. Did you know that SAMSON’s ‘Export Along Paths’ extension is designed to make this easier? Whether you’re tracking ligand pathways or studying intermediate states, this tool has you covered!
What can you achieve with ‘Export Along Paths’?
The ‘Export Along Paths’ extension allows users to:
- Export the coordinates of all atoms along a path in a system.
- Focus only on subsets of atoms, such as ligands, binding sites, or backbones, for targeted analyses.
- Save trajectories in single or multiple PDB files depending on your post-processing requirements.
With the ability to manage these details easily, modelers can generate actionable data for reaction coordinate studies or free energy profiling, reducing friction in the computational pipeline.
Step-by-Step: How to Use the ‘Export Along Paths’ Tool
Here’s a quick walkthrough of how to export atom trajectories while addressing a need frequently encountered by molecular modelers: isolating and analyzing the movement of a ligand.
Step 1: Open the Sample System
- Download a sample system by clicking on Home > Download in SAMSON.
- Paste
https://www.samson-connect.net/documents/046f1acd-c799-40f6-8185-cb4847eff795and click Download.
The sample document includes a protein structure model of Lactose permease (1PV7) and its ligand Thiodigalactosid (TDG), along with unbinding paths generated with the Ligand Path Finder extension.
Step 2: Open the Export Along Paths App
Enable the app by navigating to Home > Apps > All > Export Along Paths. You can also use the shortcut Shift + E to quickly locate the app.
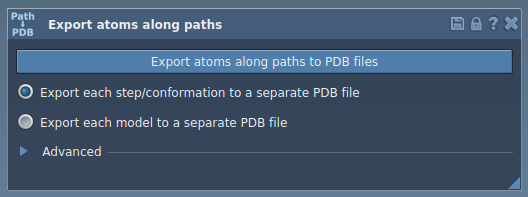
Step 3: Export Trajectories – Focusing on a Subset of Atoms
If you’re analyzing ligand motion, you might want to focus solely on the ligand:
- Expand the Advanced panel in the app interface.
- Select the ligand (TDG in this case) in the Document view.

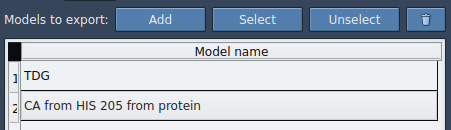
- Click Add to define the selected atoms as a model for export.
This creates a named model (e.g., ‘TDG’) in the export list. You can rename it, add additional sets of atoms, or adjust the selection at any time.
Next, select the relevant path(s) and choose an export format (single or multiple PDB files). Once ready, click Export atoms along paths to PDB files, specify a destination folder and file prefix, and you’re done!
Why Does This Matter?
Exporting trajectories has transformative implications for molecular design. You can generate input files for free energy calculations, study reaction coordinates efficiently, visualize intermediate states, or just better understand molecular dynamics—all without unnecessary manual work. With the ‘Export Along Paths’ tool, you can focus more on deriving meaningful insights from your simulations instead of losing time on tedious trajectory management.
For detailed instructions and additional tips about this feature, visit the original documentation page: Export Atom Trajectories Along Paths.
Note: SAMSON and all SAMSON Extensions are free for non-commercial use. Download SAMSON now at https://www.samson-connect.net.





