In the complex world of molecular modeling, creating realistic and biologically meaningful transition paths between different protein structures is often a challenge. If you’ve struggled with building smooth transition pathways for tasks like conformational analysis or free-energy simulations, you’re not alone. SAMSON’s ARAP Interpolator provides a fast and efficient solution to this problem.
What Is ARAP Interpolation?
The ARAP Interpolation, as implemented in SAMSON, calculates continuous structural transitions between two protein conformations. This allows molecular modelers to:
- Visualize and export intermediate conformations seamlessly.
- Generate realistic reaction coordinates for simulations like umbrella sampling.
- Align conformations with biologically meaningful geometric models.
Streamlined Workflow for Protein Interpolation
Here’s how easily you can generate transition paths with SAMSON’s ARAP Interpolator:
1. Preparing Protein Structures
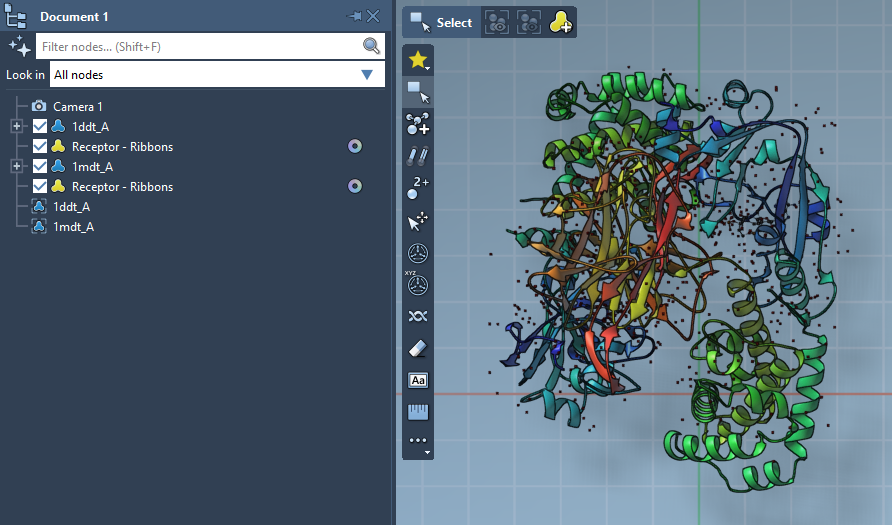
To start, use the protein structures of interest. For example, suppose you’re working with two conformations of the Diphtheria Toxin (PDB IDs 1DDT and 1MDT). Begin by fetching these structures directly in SAMSON:
- Navigate to Home > Fetch and insert the PDB IDs.
- Load the structures and remove irrelevant components such as water, ions, or chain
B.

Following this, run Home > Prepare to clean both structures. Refer to the Protein Preparation & Validation guide for additional tips.
2. Defining Conformations
Once cleaned, you’ll need to create conformations from the structures using SAMSON’s Conformation Editor:
- For
1DDT, select the structure and name its conformation1DDT A. - For
1MDT, repeat the process and name its conformation1MDT A.
These serve as the starting and goal states for interpolation.
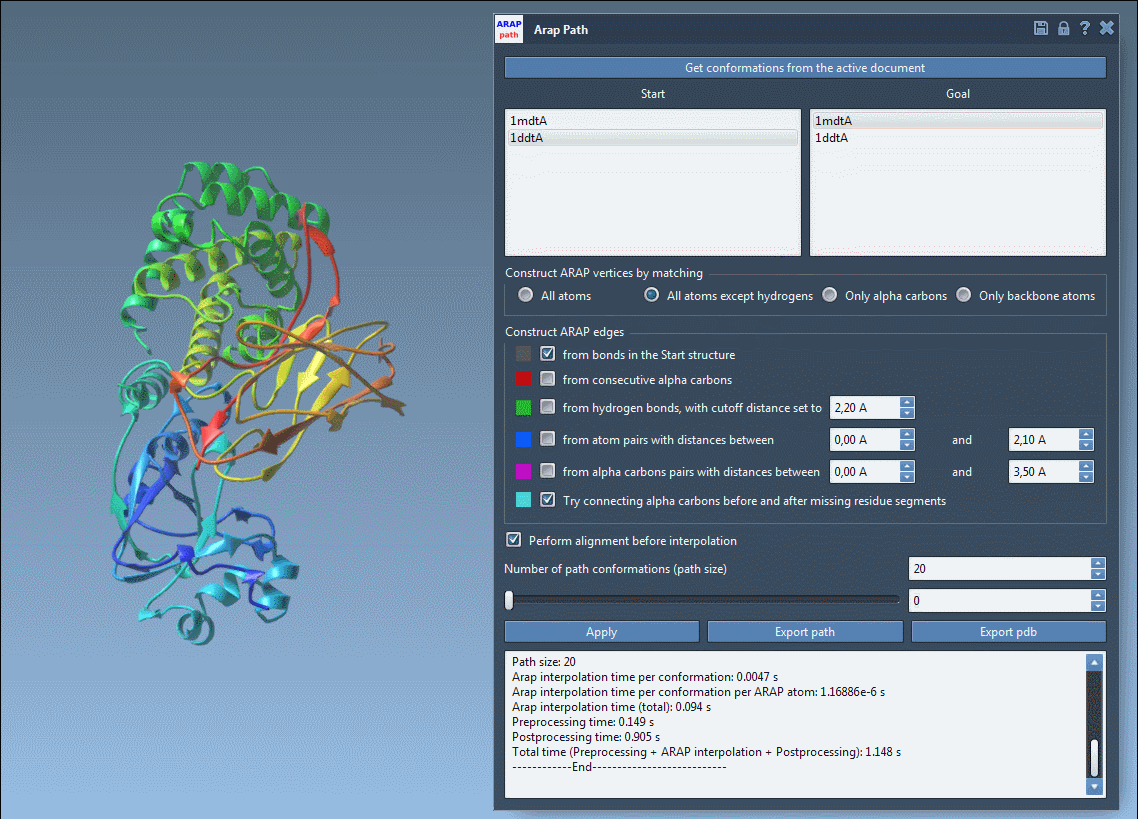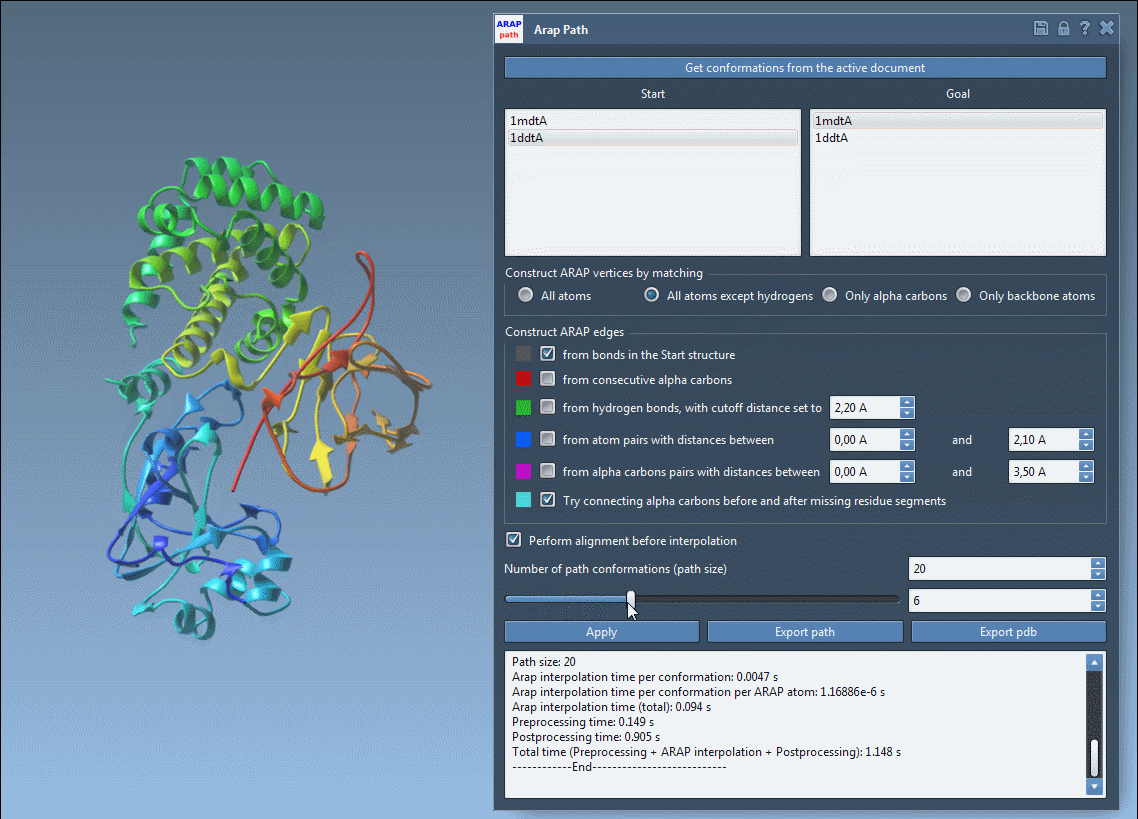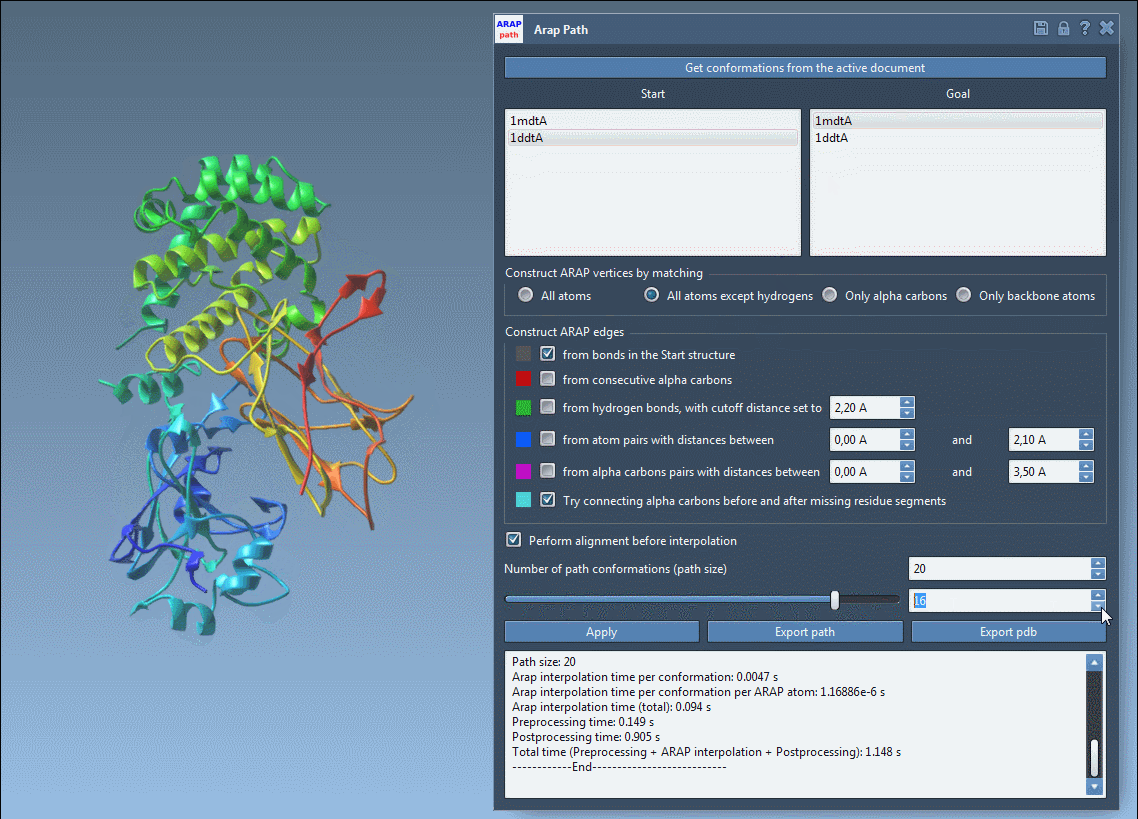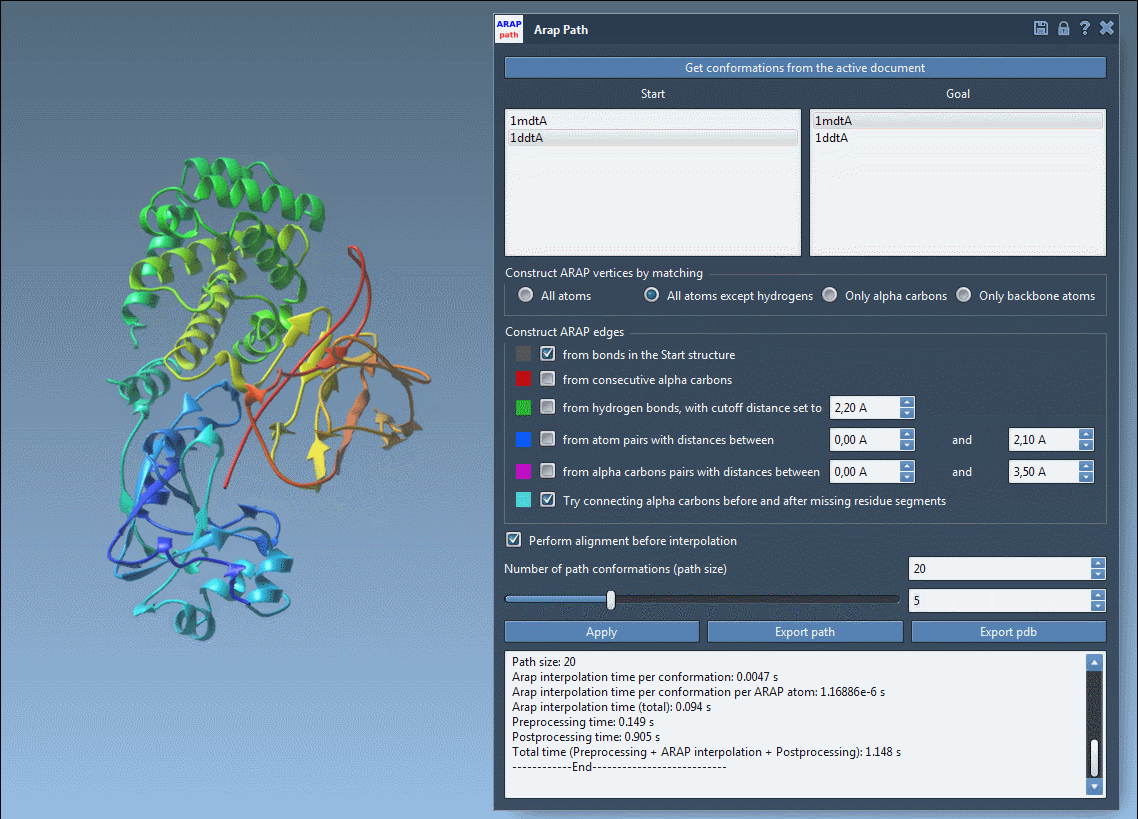
3. Interpolation Setup
Next, enter the ARAP Interpolation app from Home > Apps > Biology. Configure the parameters:
- Choose Conformations: Set
1DDT Aas the Start and1MDT Aas the Goal. - Matching Atoms: Select All except hydrogens to ensure proper vertex alignment.
- ARAP Edges: Check options like from bonds in the Start structure to maintain native connectivity.
- Preprocessing: Enable alignment and set the desired number of intermediate path conformations.
The ARAP Interpolation app offers a user-friendly interface that guides you through each step of the setup process.
4. Running and Visualizing Results
Press Run, and within seconds, your transition path will be computed. Use the integrated slider tool to examine each intermediate conformation along the path.
The computed transition path is biologically realistic and ready for further applications like free energy simulations or visualization. You can export the results in formats such as PDB for further study or analysis.
Why Choose ARAP Interpolation?
The ability to generate transition paths accurately and quickly makes the ARAP Interpolation vital for tasks like:
- Exploring dynamic structural transitions in proteins.
- Preparing accurate setups for tools like GROMACS or steered MD.
- Enhancing your understanding of conformational changes in molecular systems.
For more detailed guidance, visit the official documentation at this link.
Note: SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at this link.





