If you build custom polymers for molecular modeling, you’ve probably dealt with challenges when specifying complex monomer sequences. How do you know your defined sequence is valid? Have you correctly designated bond types between monomers? Are your naming conventions traceable?
In SAMSON’s Polymer Builder, there’s a dedicated feature to register and edit monomer sequences—a crucial step for controlling polymer architecture and minimizing errors when generating final structures. Here’s how it works, and why it might save time in your workflow.
Why Register Sequences?
When working with repeating monomer motifs, defining those as named sequences not only helps with organization but makes your polymer expressions easier to build and debug later. Instead of typing something like ABABABBA, you can just call it S1 and reuse it as needed.
How It Works
To register a new sequence:
- Click Add new sequence within the Polymer Builder interface.
- Enter your sequence using the single-letter IDs of previously registered monomers. For example:
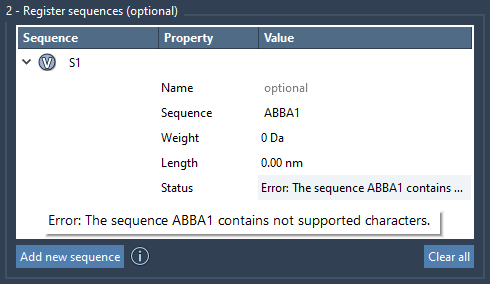
ABBA.
You can include bond types too—use = for double bonds and # for triple bonds. Here are examples:
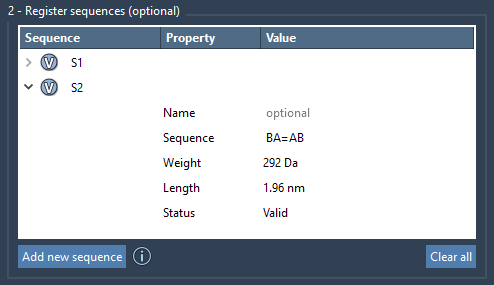
A=Bcreates a double bond between A and BA#Bcreates a triple bond between A and B
Once entered, each sequence is automatically labeled (e.g., S1, S2) and shown in a table with helpful visual cues. If there’s a syntax issue in your sequence, you’ll see an error message immediately, helping avoid silent mistakes.
Visual Feedback & Control
Next to each registered sequence is a V (Visualize) button that highlights the monomers used. This is handy when managing larger sets of sequences or when revisiting a design after time away from it.
You also have direct control over naming. Assigning a name to a sequence allows SAMSON to group its monomers under a common structural folder, which can be useful later when exporting models or running simulations in external engines.
Tidy Editing Experience
The sequence table is editable inline. You can change the order, rename sequences, or tweak bond types without redoing everything from scratch. To delete a sequence, right-click and choose Delete sequence, or use Clear all to wipe the table for a fresh start.
Bonus: Prediction
Each sequence entry shows its approximate molecular weight and length. This gives a quick sense of molecular scale before committing to polymer generation, helping you catch size mismatches early.
Conclusion
Using the sequence registration system is a good habit when designing polymers with recurring patterns or complex bonding logic. It improves traceability, reduces errors, and makes the overall design process more manageable—especially as your library of custom monomers grows.
To learn more, visit the Polymer Builder documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at www.samson-connect.net.





