One of the common pain points for medicinal and computational chemists is the time-consuming process of designing structural analogs and evaluating how small chemical modifications influence molecular interactions. Whether you’re testing binding affinity changes or investigating interaction patterns with a protein target, the manual work required can be tedious.
What if you could go from a single molecule to a full set of analogs – with 3D structures ready for analysis or docking – in just a few clicks? In this post, we’ll show how **SAMSON’s SMILES Manager** can make that possible using positional analogue scanning.
The challenge of analogue design
Traditionally, designing analogs involves using cheminformatics tools to manually search and alter fragments of molecules. Once you generate 2D structures, converting them into reliable 3D models for further analysis or docking can be another hurdle. SAMSON streamlines this with a visual, integrated approach.
Quick 3D generation from analogs
After generating analogs by defining a pattern (e.g., an aromatic carbon via the SMARTS query [cH]) and specifying what to attach or replace it with (e.g., fluorine, methyl, or nitrogen), SAMSON’s interface builds these analogs automatically. So far, so good.
But here’s where things get even more useful: you can convert all of these analogs into 3D structures in a single step. Click the Convert to 3D button, and the SMILES Manager will populate your workspace with the 3D molecular models of each analogue.
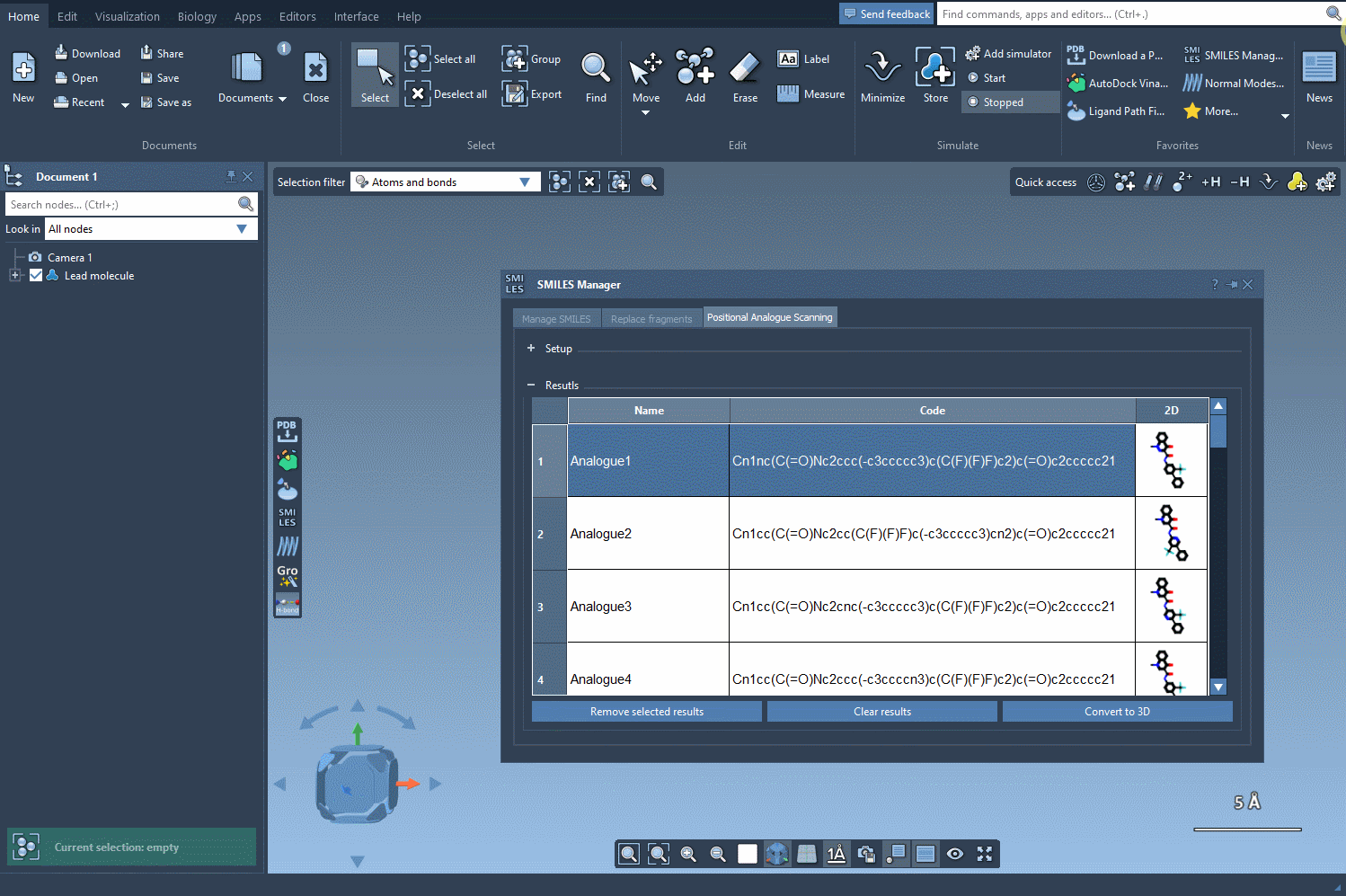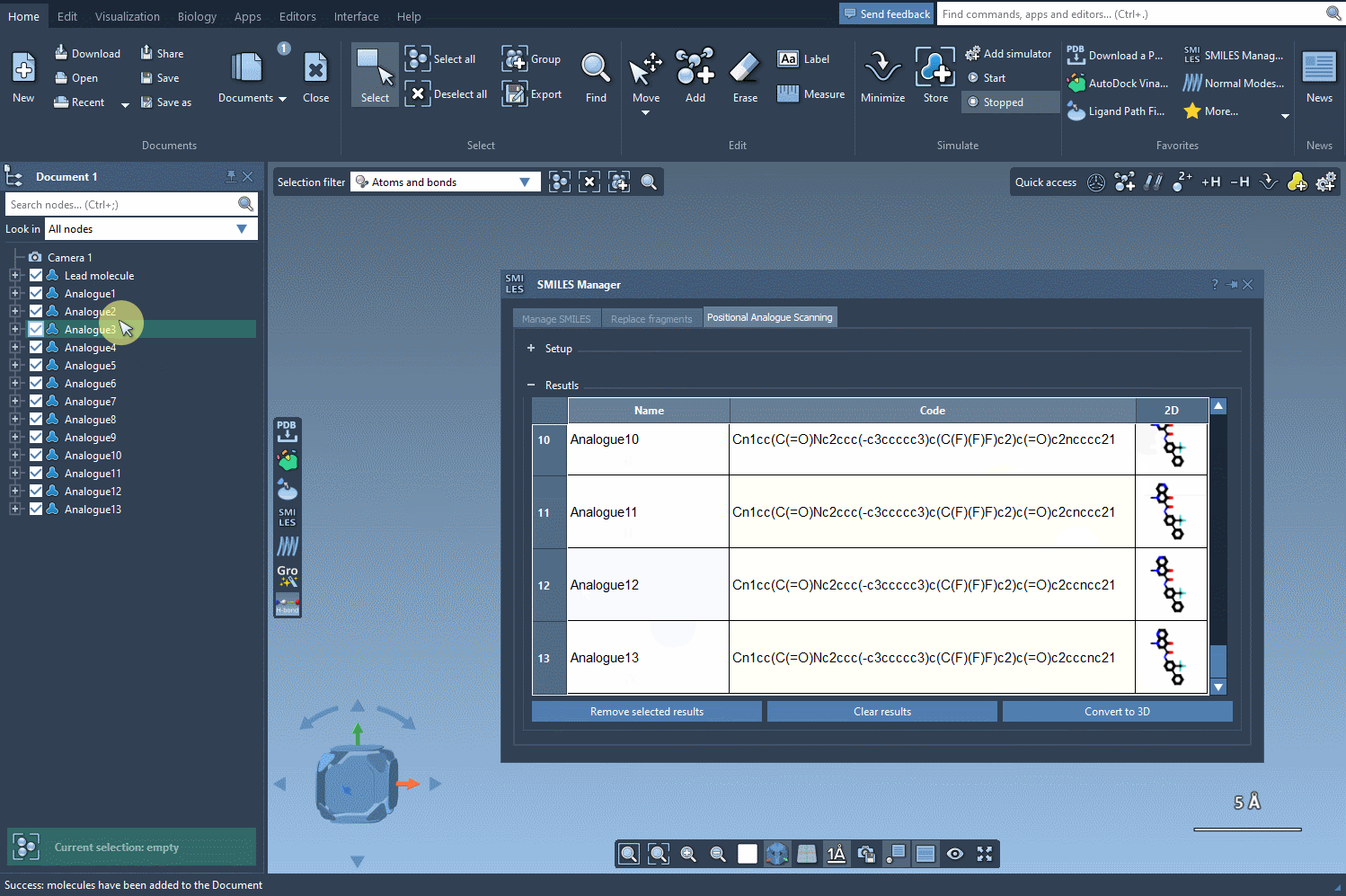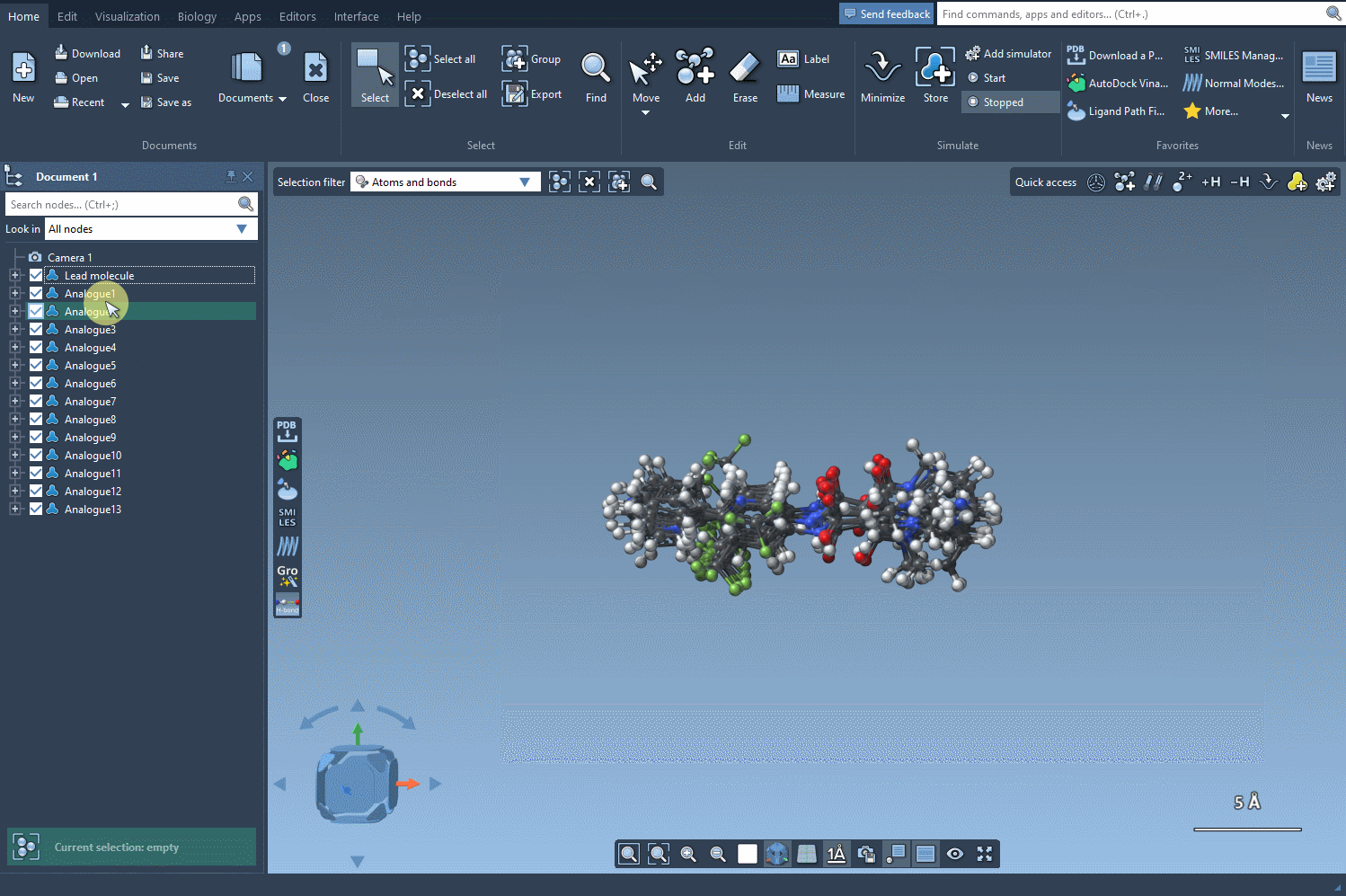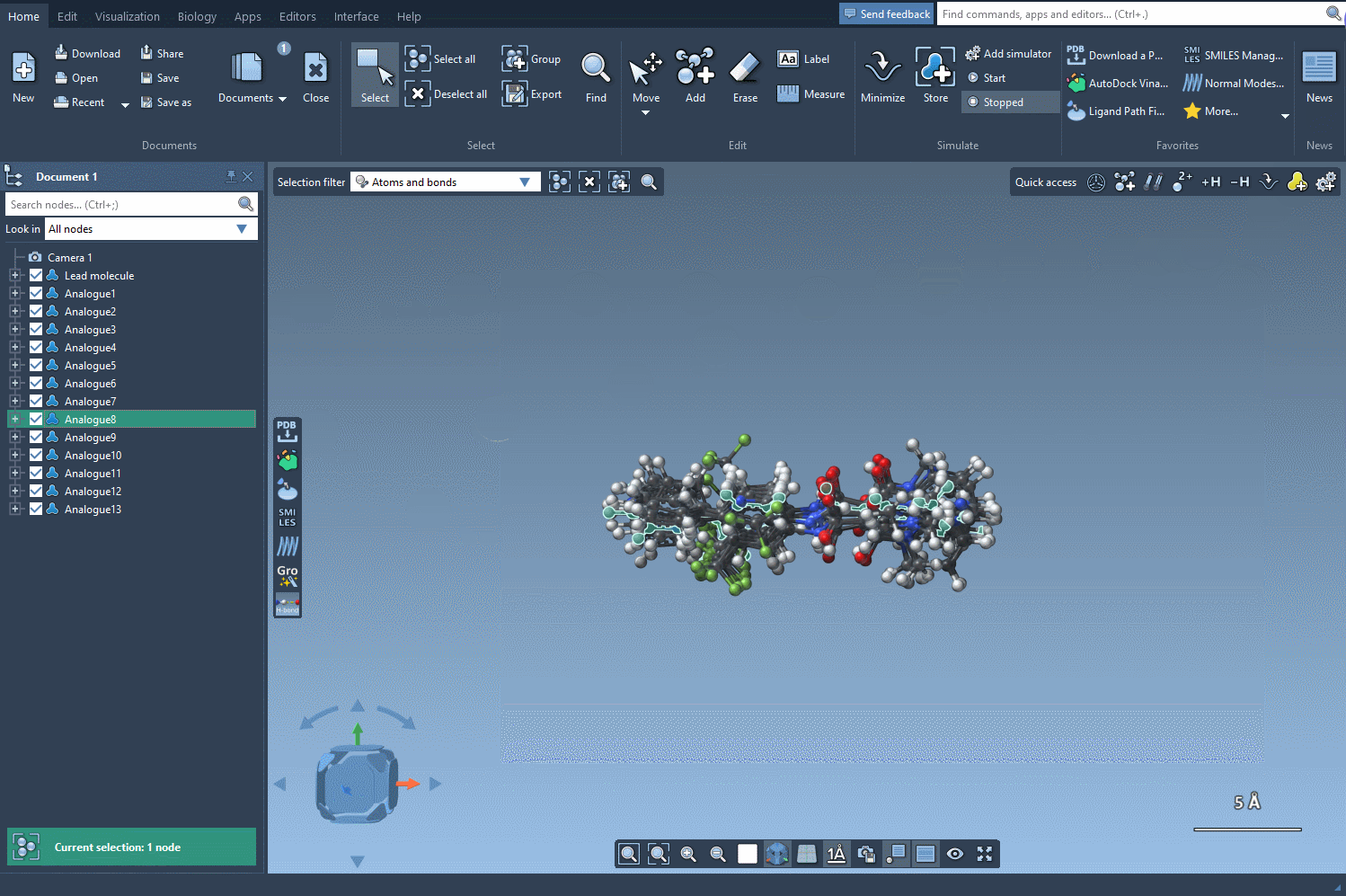
These structures are now ready for docking, interaction analysis, or any other 3D-based study you want to perform. No need to export, convert, or re-import across different tools.
What makes this useful?
This functionality is especially helpful when screening variants of a lead compound. For example, you might be testing how changing a hydrogen to a fluorine or a methyl group affects binding energy. Combining this workflow with the Autodock Vina Extended SAMSON extension, you can directly dock your analogs and identify promising candidates without leaving the platform.
It dramatically reduces the friction between idea and testing: no need to jump between cheminformatics software, visualization suites, or docking programs. Just define a pattern, generate analogs, and get 3D models – all in one place.
Small changes, big impact
Whether you’re an academic researcher or a student exploring drug design principles, this workflow enables you to focus more on thinking and less on doing repetitive work. It’s a subtle but impactful way of improving how we explore chemical space.
For more details, step-by-step instructions, and other features, see the full tutorial in the documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON here.





