If you’re a structural biologist or molecular modeler, chances are you’ve needed to visualize the conformational transition between two known protein states. Think of an important molecular mechanism—like a spike protein switching from a down to an up conformation. You might want to create an animation to present this behavior clearly to collaborators, illustrate hypotheses, or explore intermediate structures. But running full molecular dynamics simulations can be time-consuming, resource-intensive, and sometimes unnecessary.
Here’s where SAMSON’s As-Rigid-As-Possible (ARAP) Interpolation Path module comes in handy. By interpolating between two known conformations, ARAP allows you to generate a quick trajectory that visually connects the states—even if they differ in residue numbers or contain glycosylated structures like sugar-decorated viral glycoproteins.
What ARAP Interpolation Actually Does
The ARAP module doesn’t run a simulation. Instead, it generates a geometric pathway that maintains local structures while smoothly transitioning between the start and goal conformations. This makes it perfect for creating initial animations or estimating intermediate states before investing in expensive simulations.
For example, the open-to-closed transition of the SARS-CoV-2 spike protein (PDB IDs 6VXX and 6VYB) is beautifully captured in seconds using ARAP:
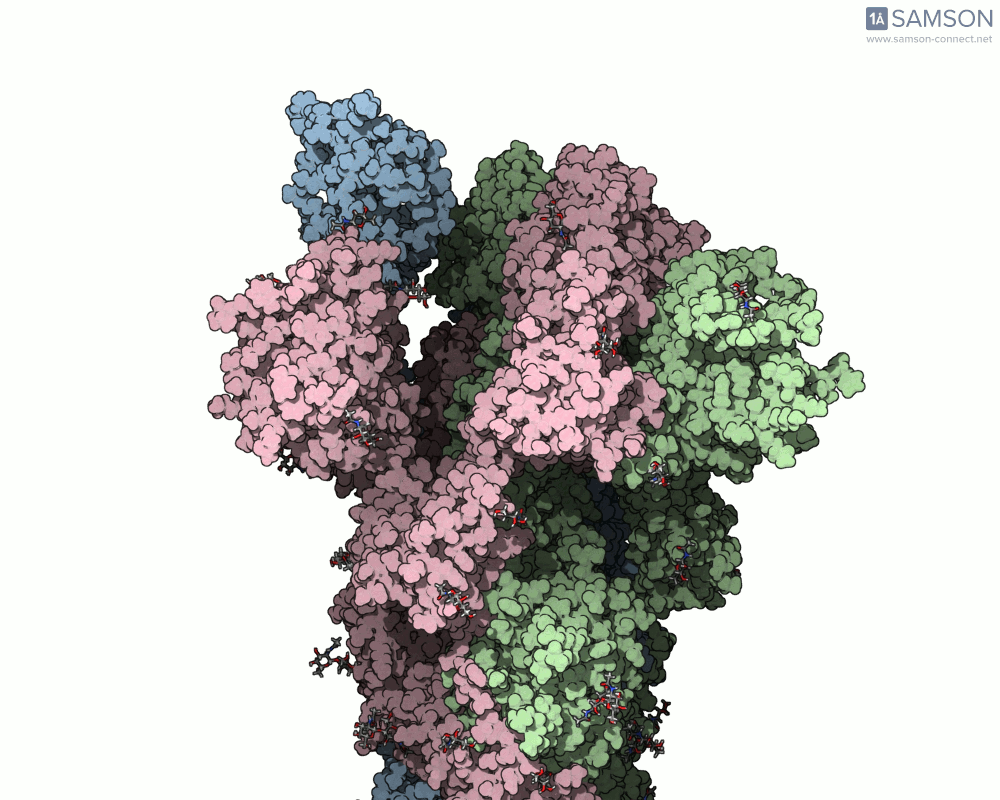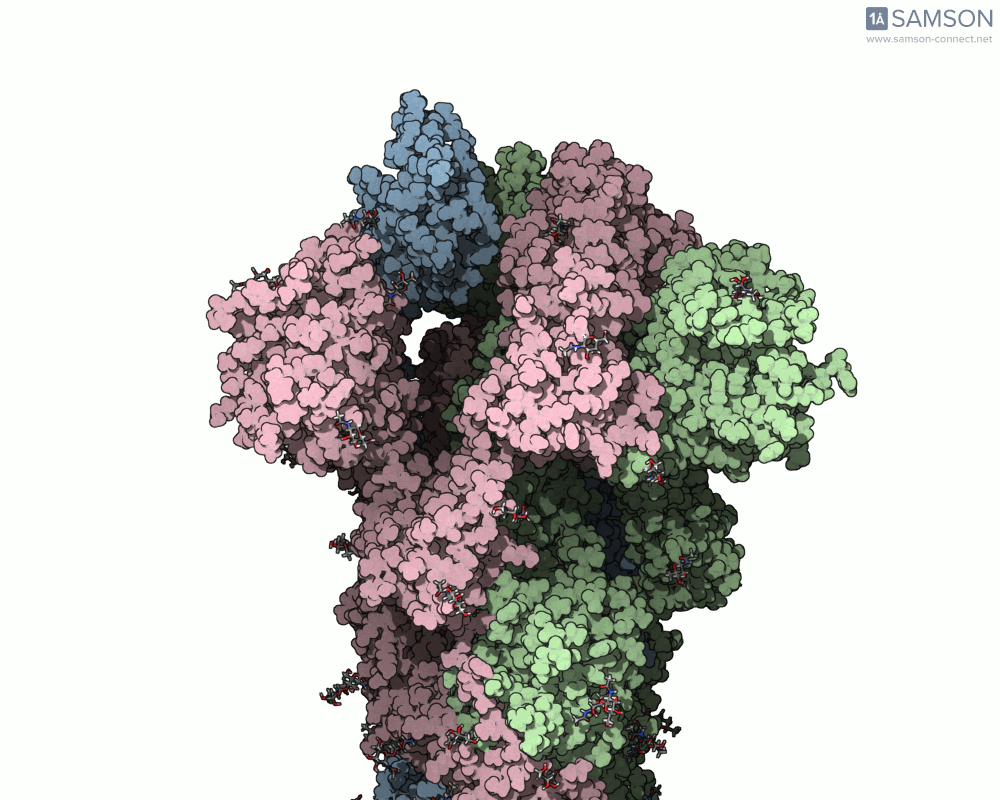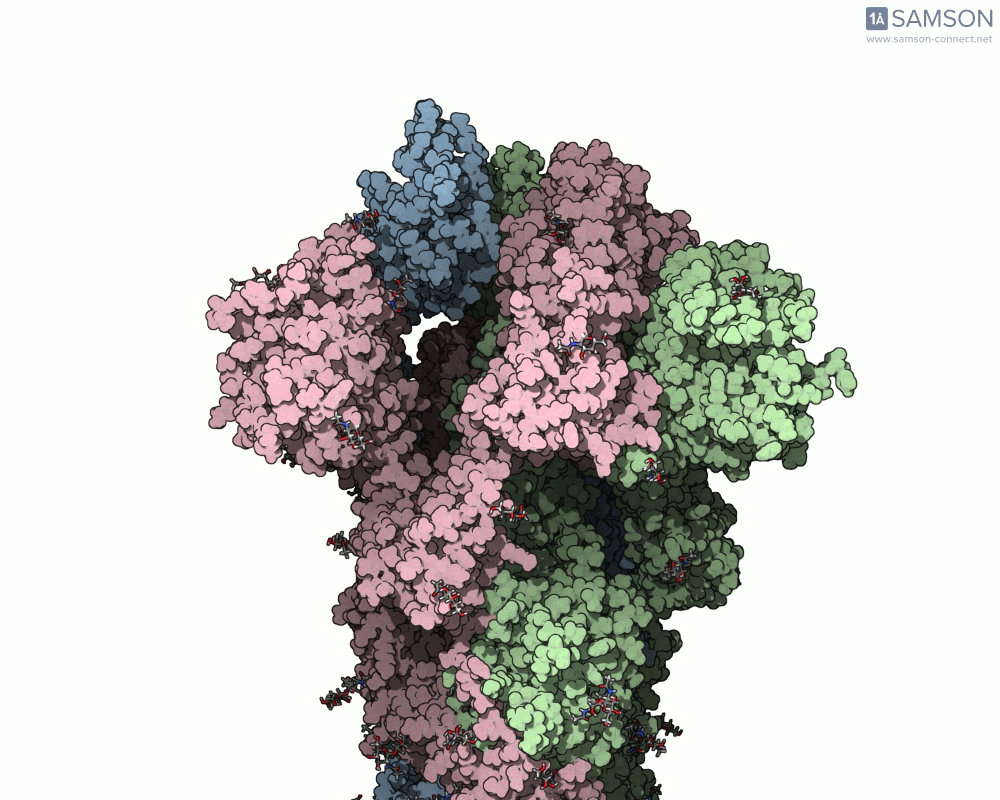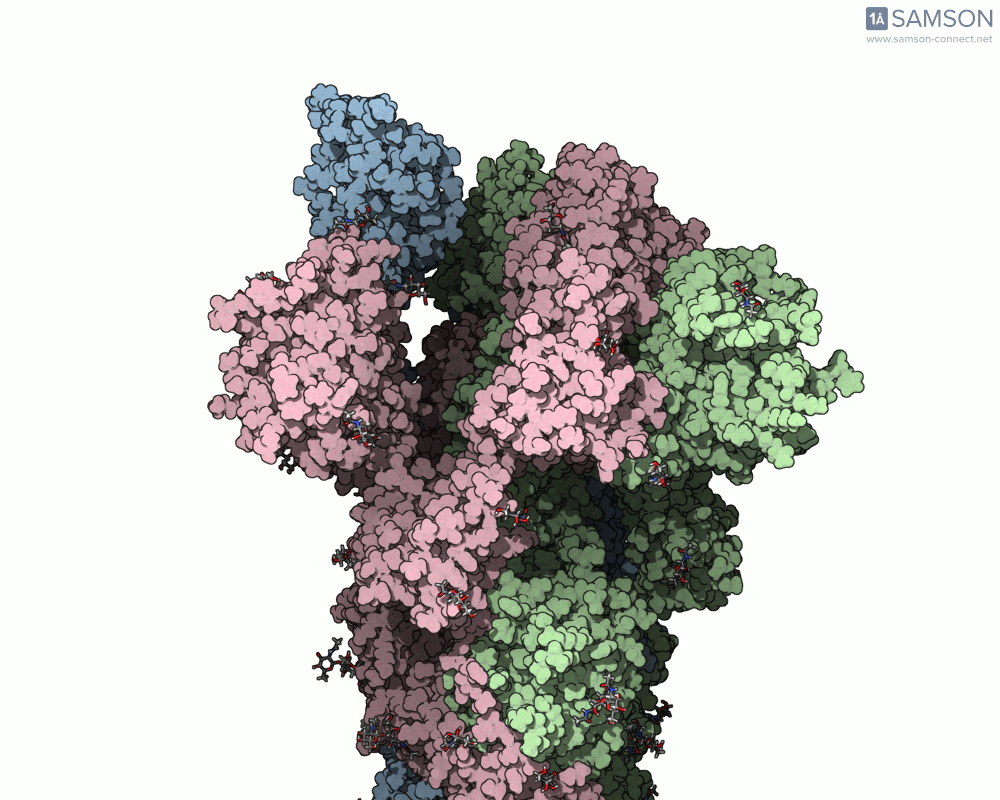
This quick generation is useful in teaching, communication, or early exploratory research where visualizing the full molecular motion helps orient design or validation steps. And because the resulting trajectory is available as a PDB file, it can be used in other tools or further refined.
Workflow at a Glance
- Prepare the two structures (open and closed) and make sure they are chemically consistent (e.g., identical bond types, hydrogens).
- Open the ARAP Interpolation Path module in SAMSON.
- Select your start (e.g., open) and target (e.g., closed) conformations.
- Run the module (it takes seconds for even large proteins).
- Visualize the trajectory or export it for additional analysis.
And if your initial structures differ somewhat in their atom or residue content, the pipeline described in the SAMSON documentation shows a solution: generate a consistent intermediate closed state from ARAP and refine it through energy minimization, before interpolating again.
Why Use ARAP?
- 🕓 Saves time—no simulations required
- 🧬 Works with large biomolecules, including glycosylated proteins
- 🖱️ Easy to use—no complex setup
- 📁 Export results to PDB or SAMSON formats for sharing or post-processing
Whether you’re illustrating protein function or preparing for more in-depth computations, ARAP provides a fast and intuitive way to animate conformational changes.
To learn more and follow the full walkthrough, visit the original SAMSON tutorial page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





