If you’ve ever needed to extract the trajectory of a ligand along an unbinding path for further analysis, you’re not alone. For molecular modelers working on free energy calculations, reaction coordinate generation, or enhanced sampling setups, being able to export atomic positions along a specific pathway is a recurring task.
This is where the Export Along Paths extension in SAMSON can save time and reduce complexity. In this post, we’ll walk through how to export only a subset of atoms—for example, a ligand—along a predefined path. This keeps your output lean and focused.
Why export only a subset?
Sometimes, especially in binding or unbinding studies, you’re only interested in the ligand’s behavior—not the entire system. Exporting just the ligand trajectory helps set up reaction coordinate windows, generate input for umbrella sampling, or visualize ligand motion without unnecessary structural data.
Start by installing the extension
- Log into SAMSON Connect.
- Visit the Export Along Paths page and click Add.
- Restart SAMSON. The tool is now available in your interface.
Load the sample system (optional but useful)
You can try this with our sample system: a structural model of Lactose permease with its ligand Thiodigalactosid (TDG) and pre-calculated unbinding paths.

Exporting a subset: just the ligand
- Launch the Export Along Paths app via Home > Apps > All > Export Along Paths.
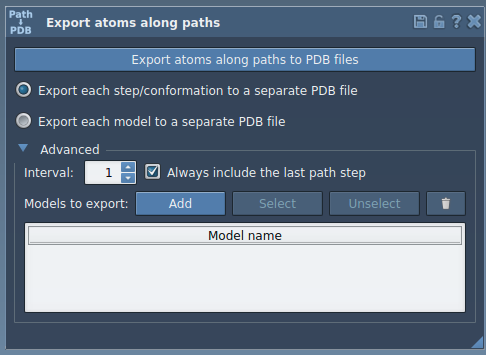
- Expand the Advanced section in the app interface.
- In the Document view, select the ligand (e.g., TDG).

- Click Add in the app to define the selected atoms as a model for export.
Before you export
- The new model (TDG, in this case) appears in a table. You can rename it by double-clicking on its name.
- To inspect atoms included in each model, click Select. To redefine them, use the Reset button.

Exporting
- Select the relevant path(s) in the Document view.
- Choose the export format (one or multiple PDB files).
- Click Export atoms along paths to PDB files.
You’ll be prompted to choose a folder and file name prefix.
Use cases where this really helps
- Creating reaction coordinate files for PMF calculations
- Preparing input for umbrella sampling simulations
- Visualizing ligand unbinding/entry independently of receptor movements
This kind of selective export can help keep datasets compact and workflows more manageable.
To learn more and explore related workflows, check the full documentation page: https://documentation.samson-connect.net/tutorials/export-along-path/export-atoms-trajectories-along-paths/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON here.





