Author: OneAngstrom
Avoid Boundary Artifacts in Molecular Dynamics: How to Set Up the Simulation Box for COM Pulling
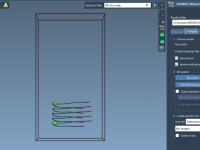
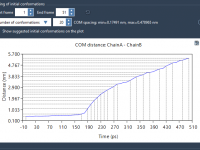
If you’ve ever run molecular dynamics (MD) simulations with pulling forces—especially center-of-mass (COM) pulling—you’ve likely encountered artifacts caused by poorly defined simulation boxes. These artifacts can significantly compromise the realism of your results. In this post, we’ll explore how you…
Tired of Compatibility Issues? Here Are the File Formats SAMSON Supports
Filtering Molecular Structures with NSL in SAMSON’s Document View
How to Quickly Design and Reuse Monomer Sequences in SAMSON
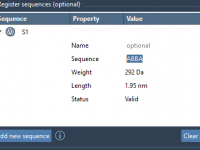
Designing custom polymers often requires handling complex monomer sequences—whether you’re modeling repetitive structures, testing new arrangements, or building realistic polymers for simulations. One time-consuming step many molecular designers face is the repetitive reconstruction of commonly used monomer sequences. With SAMSON’s…
Freeing Ligands: A Simple Way to Animate Undocking in Molecular Presentations
Modifying GROMACS MDP Parameters in SAMSON Without the Hassle
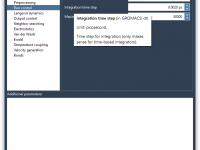
For molecular modelers using GROMACS, adjusting simulation parameters typically involves diving into .mdp files and memorizing configurations. This can become time-consuming and error-prone—particularly when repeating simulations with slight variations or collaborating across teams. The GROMACS Wizard in SAMSON offers an…
Launching GROMACS Batch Simulations for Multiple Molecular Conformations
How to Create Custom Index Groups for GROMACS Simulations without Writing Selection Scripts
Custom index groups can be extremely helpful in molecular dynamics simulations, especially when performing detailed analyses or steering parts of your system through advanced scenarios like umbrella sampling or pulling simulations. However, defining these groups using command-line tools and selection…