Author: OneAngstrom
A practical look at selecting node groups in SAMSON
Avoid This Common Pitfall When Preparing Proteins for Conformational Analysis
A quick way to find a color scheme that works for your molecular models
Choosing the right color palette is essential when building or analyzing molecular models, especially when attempting to convey complex information clearly. Whether you’re preparing figures for a scientific publication, explaining results during a presentation, or exploring molecular systems visually during…
Keeping Your Eye on the Action: Dynamic Focus with ‘Look at Atoms’ in SAMSON
Avoiding Errors in NPT Equilibration: How to Use the Right Input in GROMACS Wizard
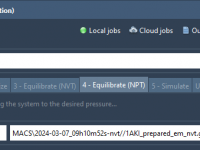
Molecular dynamics (MD) simulations often involve long workflows composed of successive stages: minimization, temperature equilibration (NVT), pressure equilibration (NPT), production runs, and post-analysis. Each step builds on the previous one. However, a frequent frustration among modelers—especially those new to MD…
Why Can’t I Just Export to PDB? Tips for Managing File Formats in Molecular Modeling
When Pressure Matters: Stabilizing System Density with NPT Equilibration in SAMSON
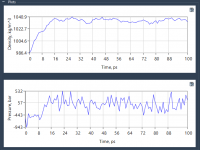
One common challenge molecular modelers face lies in achieving a stable simulation environment after initial energy minimization and temperature equilibration. Even with a minimized system at the right temperature, unstable density can still derail longer molecular dynamics (MD) simulations. Fortunately,…
Speed Up Your Molecular Modeling: Filter Cameras in SAMSON with Precision
When working on complex molecular scenes in SAMSON, it’s easy to accumulate multiple camera nodes—each representing different viewpoints, lighting setups, or visual documentation stages. But managing them efficiently can become challenging, especially when quickly switching perspectives or automating workflows. That’s…
From PDB to Motion: Visualizing Spike Protein Transitions in SAMSON
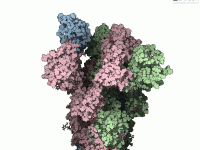
Generating biologically meaningful motion pathways between molecular conformations—especially for large biomolecules—is a common challenge in structural biology and molecular modeling. When the structures differ in atom count or residue indexing, things can get especially tricky. If you’ve ever attempted to…