Author: OneAngstrom
When Temperature Matters: Navigating NVT Equilibration in GROMACS Wizard
Installing and Editing Local Python Packages Inside SAMSON
Many molecular modelers and computational chemists have their own Python packages—for custom simulations, analysis pipelines, or even neural networks for property prediction. But using them often means jumping between development environments, terminal sessions, and structured molecule viewers. What if everything…
Quickly See or Hide Molecular Animation Nodes with SAMSON’s Visibility Attributes
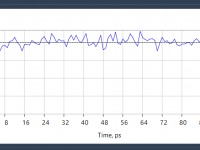
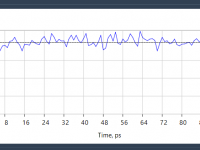
Quickly Stabilize Temperature in Molecular Simulations with NVT Equilibration in SAMSON
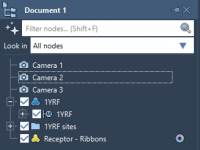
A Better Way to Navigate Your Molecular Scenes: Using Multiple Cameras in SAMSON
Quickly Target Node Groups in SAMSON with the Node Specification Language
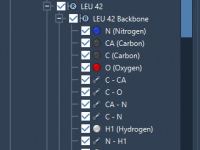
Modifying GROMACS parameters in SAMSON without breaking your workflow
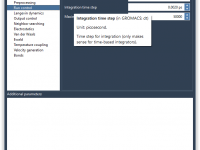
One of the recurring pain points for molecular modelers using GROMACS is managing and fine-tuning molecular dynamics parameters across simulations. Whether you’re running energy minimizations or full production MD simulations, adjusting .mdp files efficiently without making mistakes or interrupting your…