Author: OneAngstrom
Controlling Molecular Visibility Over Time with the Hide Animation in SAMSON
Why Your Molecules Look Flat (and How to Fix It)
Efficiently Zoom Into Molecular Details Without Changing Your Scene
When Bonds Break: Modeling Topological Changes in Real Time
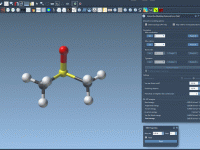
Designing molecular systems often involves experimenting with structures: forming bonds, breaking them, tweaking atom types. For many molecular modelers, these changes have traditionally required discrete steps such as redefining topology, restarting simulations, and recalculating parameters—interrupting the creative or scientific flow.…
Making Molecular Models Gradually Appear in Your Presentations
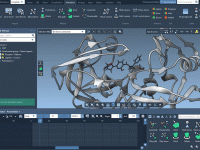
Designing clear and engaging molecular presentations can be tricky—especially when it comes to showing complex molecular structures without overwhelming the viewer. One recurring challenge for molecular modelers, educators, and students is how to highlight parts of a molecular system progressively,…
How Sampling Box Setup Affects Ligand Unbinding Pathways
Saving Time with AlphaFold-2: Protein Prediction in SAMSON, Step by Step
Quickly Identifying Files of Interest in Molecular Projects
Why Your Molecular Animation Suddenly Jumps—And How to Fix It
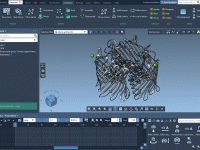
Molecular visualizations can be powerful storytelling tools. Whether you’re preparing a presentation, a scientific movie, or just documenting your molecular model’s behavior over time, a smooth camera trajectory is crucial. However, many molecular modelers have encountered this frustrating issue: suddenly,…