Author: OneAngstrom
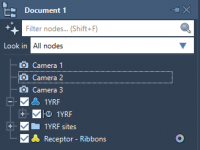
Why Molecular Modelers Should Start Using Multiple Cameras
Quickly identify specific bond types in your molecular models
Making Molecules Rock: A Simple Way to Add Movement to Your Models
Quickly Select Backbone Groups with Specific Atomic Composition in SAMSON
In molecular modeling projects involving large biomolecular systems, tasks such as identifying backbone structural groups with specific atomic counts or charge characteristics can be time-consuming. Whether you’re preparing a dataset, analyzing systems, or building coarse-grained models, manually filtering and inspecting…
When and Why You Should Minimize Ligands Before Docking
Showing Disappearance in Molecular Models: A Closer Look at the ‘Conceal Atoms’ Animation
Orbit Camera: A Simple Way to Get Stunning Molecular Visuals
Bringing Your Molecular Tools into SAMSON
Many molecular modelers face a familiar challenge: you’ve already developed tools or use third-party software for simulations, docking, or visualization — but managing interoperability becomes time-consuming and error-prone. If you’re switching between tools or manually parsing results across platforms, you’re…