If you’ve ever performed Potential of Mean Force (PMF) calculations using GROMACS, you probably remember the long list of scripts and careful file organization required to properly apply the Weighted Histogram Analysis Method (WHAM). Piecing together dozens of outputs from umbrella sampling windows, aligning data formats, and plotting can be time-consuming—and prone to mistakes.
Thankfully, the GROMACS Wizard in SAMSON simplifies this process dramatically by offering a visual, interactive way to perform WHAM-based PMF analysis. In this post, we’ll walk you through how this works and how it can help eliminate a recurring pain point in molecular modeling workflows.
Why PMF Matters – and Why It’s Complicated
PMF profiles provide insight into the free energy landscape along a chosen reaction coordinate, allowing researchers to study molecular mechanisms like ligand binding, ion permeation, or conformational transitions. However, the steps required to conduct this analysis manually are often tedious:
- Running many umbrella sampling simulations with consistent parameters
- Caching output data in structured folders
- Formatting and postprocessing trajectory data
- Applying WHAM via command line
- Plotting results using external tools
Each of these steps requires care, and even small missteps in file handling or parameter settings can compromise the end result.
Streamlining PMF with GROMACS Wizard
With GROMACS Wizard in SAMSON, you can load your umbrella sampling results and perform WHAM-based analysis directly via a graphical user interface. Here’s how it works:
After completing umbrella sampling simulations—either manually or using GROMACS Wizard’s Batch Computation tool—you can switch to the WHAM Analysis tab:
![]() Click the auto-fill button to automatically detect the path where your simulation outputs are stored. All umbrella sampling folders should be numbered and should contain simulation results using the same reaction coordinate.
Click the auto-fill button to automatically detect the path where your simulation outputs are stored. All umbrella sampling folders should be numbered and should contain simulation results using the same reaction coordinate.
Once the project path is loaded, GROMACS Wizard reads in the available information – such as reaction coordinates, simulation time, and temperature – and offers a drop-down for selecting the reaction coordinate to analyze. You can also fine-tune the analysis using custom bounds, time ranges, and units.
Clicking Compute launches the WHAM calculation. The results include both the PMF profile and the coverage histogram:
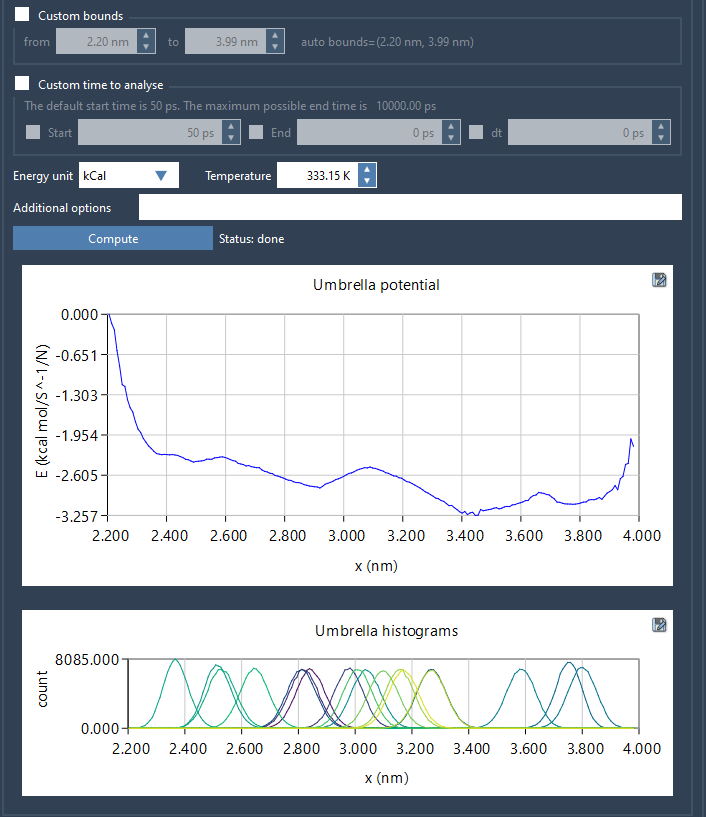
The PMF plot reveals the approximate free energy surface along your reaction coordinate, while the histogram shows how well your sampling covered that space. If the histogram reveals sparse areas, you’ll know exactly where to add more sampling before rerunning WHAM.
Reusability and Data Management
Computed results are automatically stored in a wham_results subfolder in your project. This data is reused when you switch between reaction coordinates or explore different parameters, allowing for efficient iteration without redoing calculations.
Summary
If you’re working with umbrella sampling data and want a more intuitive way to perform PMF analysis, the GROMACS Wizard in SAMSON offers a user-friendly solution. It takes care of folder structuring, parameter consistency, WHAM computation, and plotting—all from one place—so you can focus more on interpreting results than on managing files and scripts.
Learn more about PMF analysis in GROMACS Wizard by visiting the official documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





