When computing the Potential of Mean Force (PMF) in molecular simulations, there’s a crucial detail that can often be overlooked: the quality of sampling along the reaction coordinate. Poor sampling can lead to misleading energy profiles, potentially invalidating the conclusions of your modeling. This is why visualizing the histogram in PMF computations using the Weighted Histogram Analysis Method (WHAM) is so important. Fortunately, the GROMACS Wizard in SAMSON makes this easier by integrating histogram diagnostics directly into the workflow.
Let’s break it down for anyone performing umbrella sampling simulations:
What’s the issue?
When umbrella sampling is used to create biased simulations along a reaction coordinate, the quality of the final PMF depends heavily on how well the states (or windows) cover the entire reaction coordinate space. If some parts are under-sampled, WHAM cannot reliably reconstruct the free energy profile, potentially producing artifacts or underestimating barriers.
How GROMACS Wizard helps
After you run your umbrella sampling simulations — either manually or via batch computation — you can switch to the WHAM Analysis tab in the GROMACS Wizard. Here’s where things get interesting.
When you select your reaction coordinate and hit Compute, two plots are generated:
- PMF curve — shows the effective free energy landscape.
- Histogram — reveals how well each segment of the reaction coordinate has been sampled.
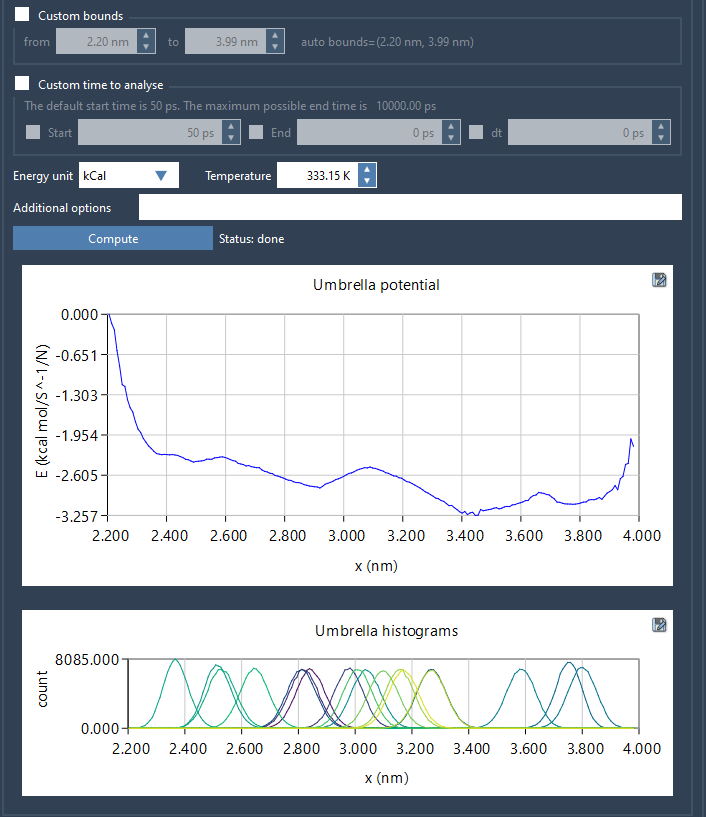
The histogram is not just a side plot — it’s your diagnostic tool. If you see gaps or low counts in parts of the reaction coordinate range, it’s a sign that the sampling there might be insufficient. This allows you to decide whether to:
- Add more windows to improve coverage
- Extend simulation times in existing windows
- Adjust force constants to better overlap between neighboring windows
Input folder structure matters
If you’re coming from an umbrella sampling run, the GROMACS Wizard helps you by auto-filling the project path with a single click. The folder must contain numbered subfolders (e.g., 000, 001, etc.), each representing a window with its own simulation data.
Built-in storage and reuse of results
What’s helpful is that once a PMF is computed for a certain reaction coordinate and its options (bounds, time window), the results are cached in the wham_results folder. So, switching between reaction coordinates or trying different parameters doesn’t mean recomputing everything from scratch — a good time-saver.
Takeaway
Whether you are studying ion channels, host-guest systems, or protein-ligand dissociation pathways, always check the histogram when computing PMFs. It’s an essential part of WHAM analysis — and with GROMACS Wizard’s integrated plots, understanding where more work is needed becomes much easier.
To learn more, visit the official documentation page: https://documentation.samson-connect.net/tutorials/gromacs-wizard/pmf-analysis/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





