One persistent challenge for molecular modelers is the need to rebuild molecular topologies every time structural changes occur. Whether you’re exploring reaction pathways, molecular docking, or simply correcting a bond type, switching between building mode and simulation mode can disrupt your workflow.
What if you could modify molecules — break bonds, change bond orders, and create new connections — while simulating, without breaking continuity or needing to halt and reconfigure the model?
This is where the Interactive Modeling Universal Force Field (IM-UFF) in SAMSON offers a practical solution.
Smooth Topological Transitions, Real-Time Control
The IM-UFF interaction model enables users to perform simulations in real time, even as the underlying molecular topology dynamically changes. With IM-UFF, you can:
- Break covalent bonds by dragging atoms away from their neighbors 🧪
- Create new covalent bonds by moving atoms close together
- Let bond order and atom typization adjust automatically based on distances
Most force fields assume a static topology, making them unsuitable for exploratory modeling. The ability to switch topology on-the-fly is especially useful in conceptual design, education, and early-stage hypothesis building where structure exploration matters more than computing energetics with chemical precision.
How to Start an IM-UFF Simulation
Here’s how to get going with dynamic interactive modeling in SAMSON using IM-UFF:
- Open a molecular system document in SAMSON.
- Go to Edit > Simulate > Add simulator, or use the shortcut:
Ctrl + Shift + M(Windows/Linux) orCmd + Shift + M(macOS). - Choose Interactive Modeling Universal Force Field as your interaction model.
- Select a state updater, such as FIRE.
Then, start the simulation with Edit > Simulate > Start. Make sure the following two options are unchecked in the IM-UFF parameter window:
- Static topology (UFF only)
- Keep vdW for manipulated (optional, based on your manipulation needs)
Watch the Process in Action
Here’s an example of bond breaking and formation happening dynamically as a user drags atoms with the mouse:
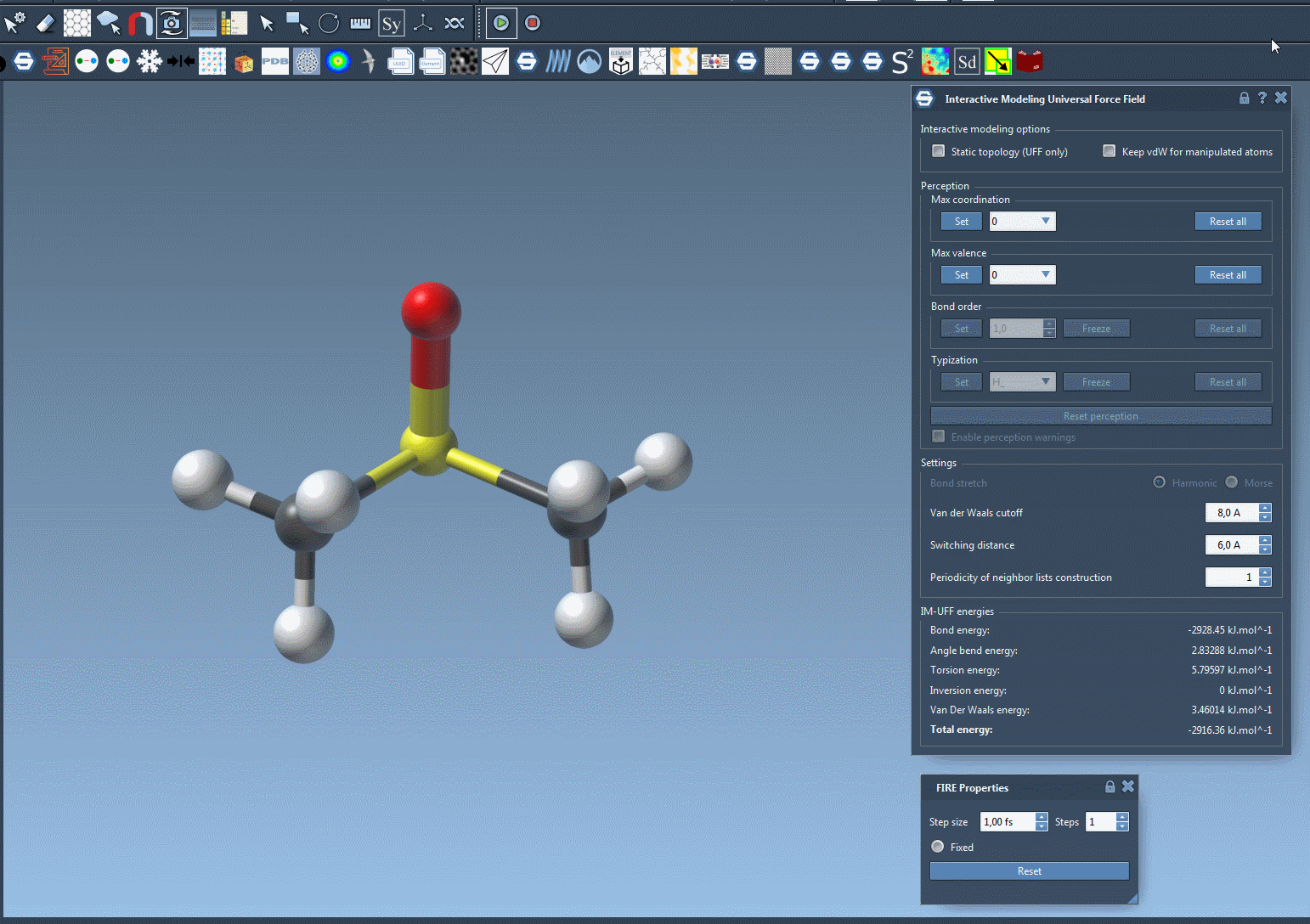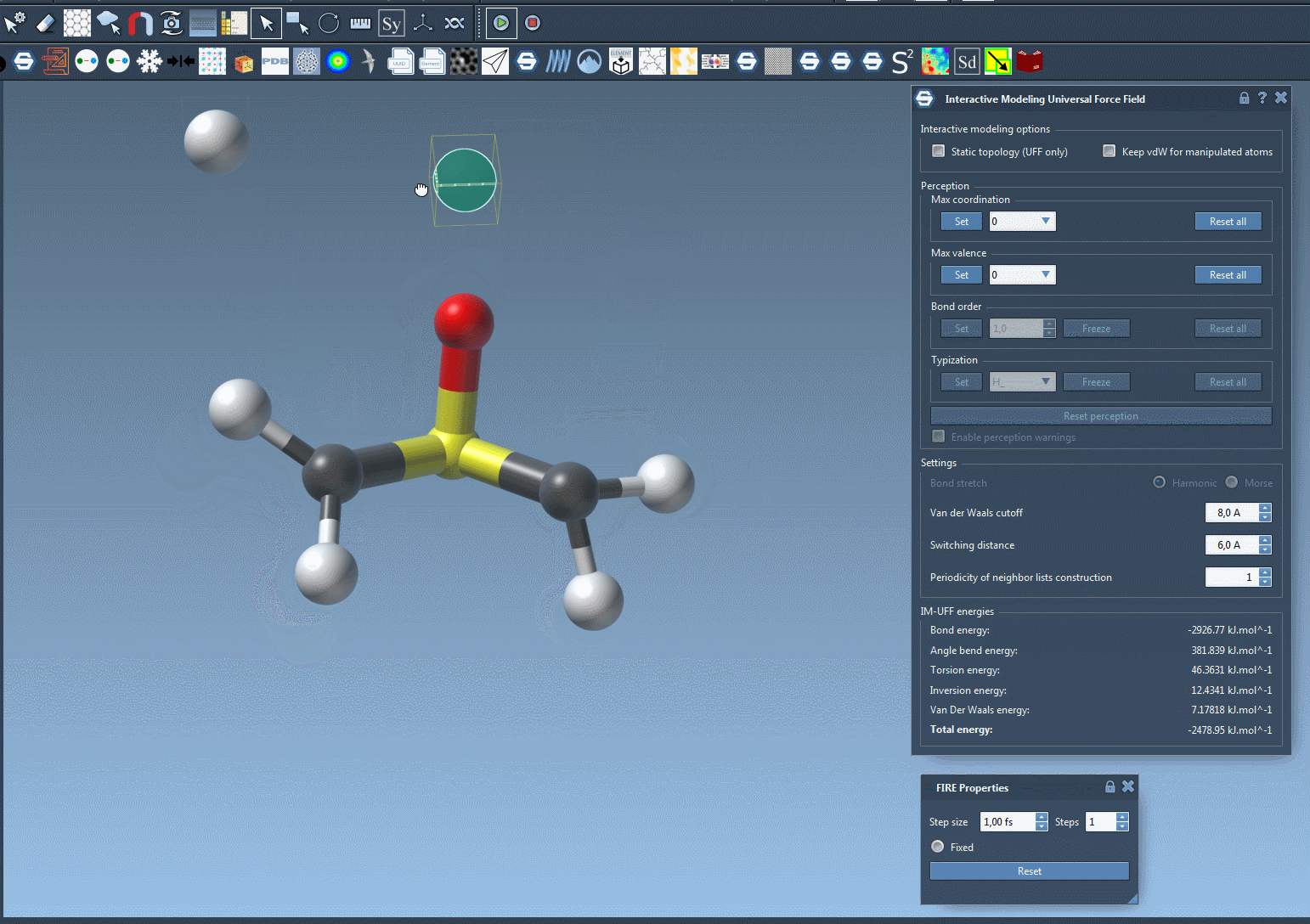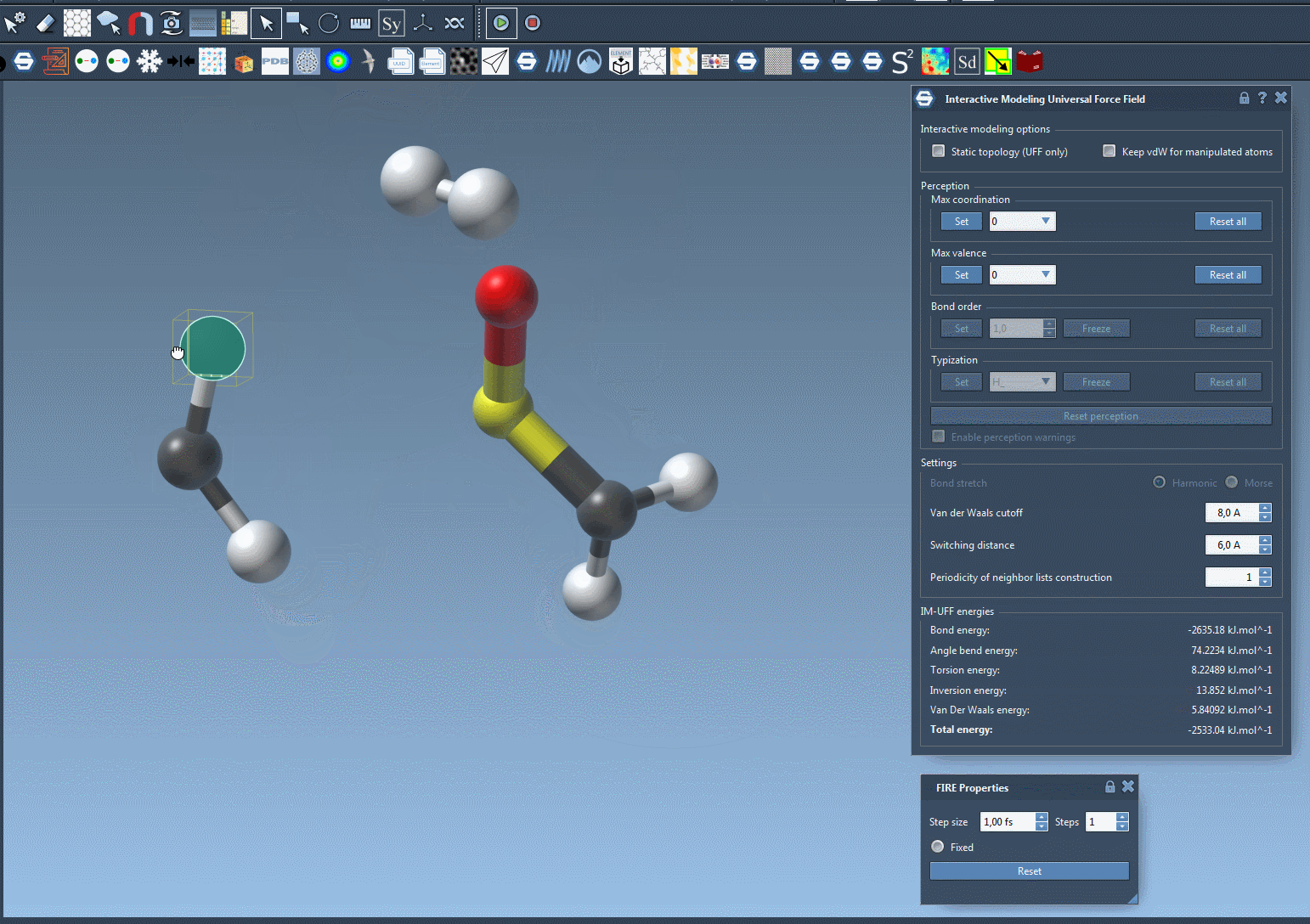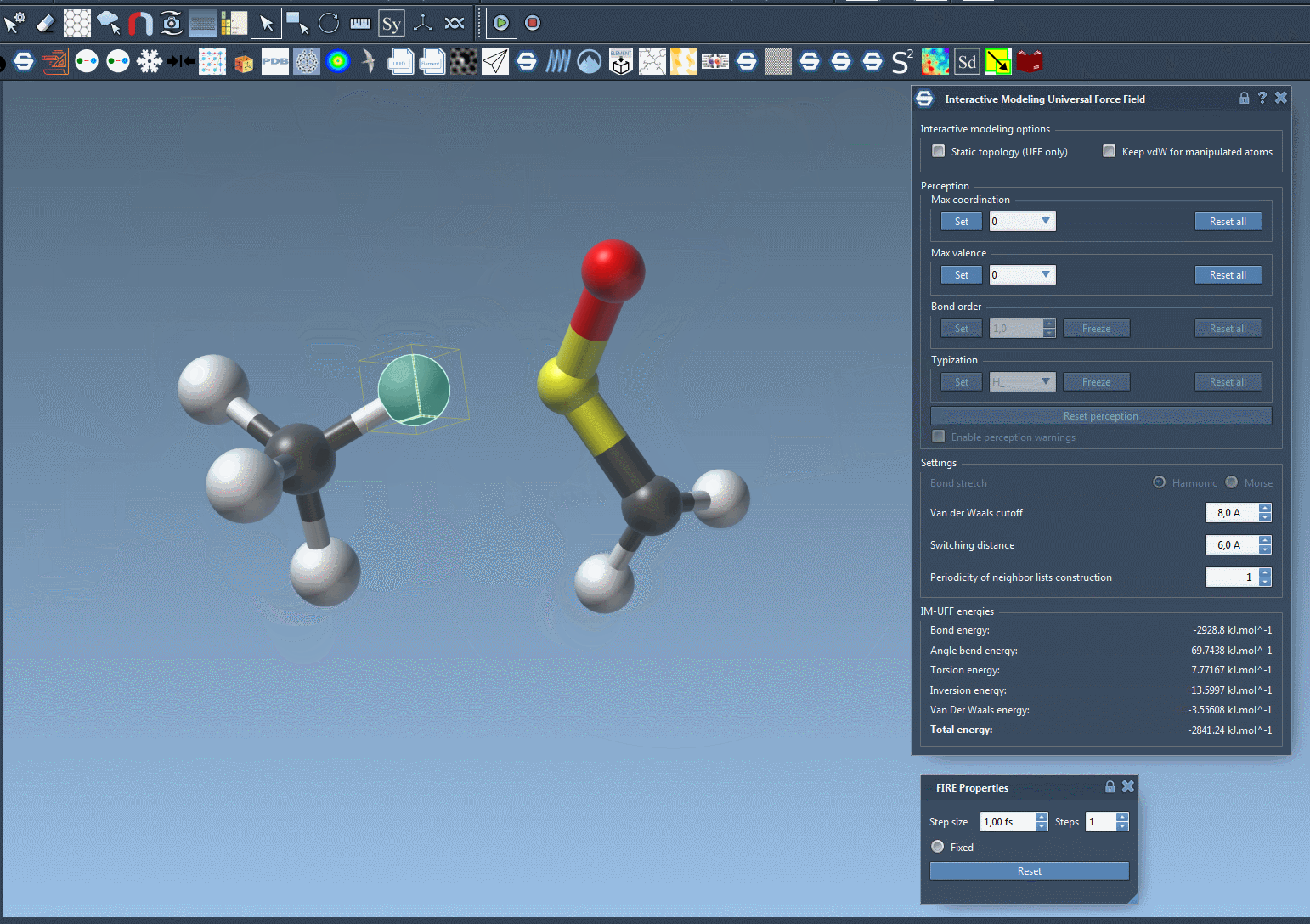
You’ll observe the total energy of the system at the bottom of the parameter window. Displacing atoms slightly causes local rearrangements, while more drastic displacements lead to bond breaking. Bring atoms close enough, and new bonds may form.
Why This Helps
This interactive modeling mode can be especially helpful when:
- Experimenting with chemical reactions
- Trying out structural modifications quickly
- Teaching molecular modeling and explaining topology changes
- Prototyping new molecular structures before performing high-accuracy quantum calculations
Because the changes are handled by the force field itself, you don’t need to pause and update your topology manually. This can save hours during early-stage molecular design studies.
To learn more, visit the full documentation: IM-UFF Tutorial.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





