Setting up simulations of membrane proteins often starts with a significant bottleneck: building a reliable lipid layer around a protein. This step is critical for many molecular modeling projects, from studying membrane protein dynamics to simulating lipid-mediated interactions. Yet, the process usually requires either scripting efforts or dependence on complex third-party tools.
The Molecular Box Builder in SAMSON offers a simple approach to build lipid layers around proteins. It handles orientation, box definition, and packing—graphically and intuitively—so you can move past the tedious setup and get to simulation faster.
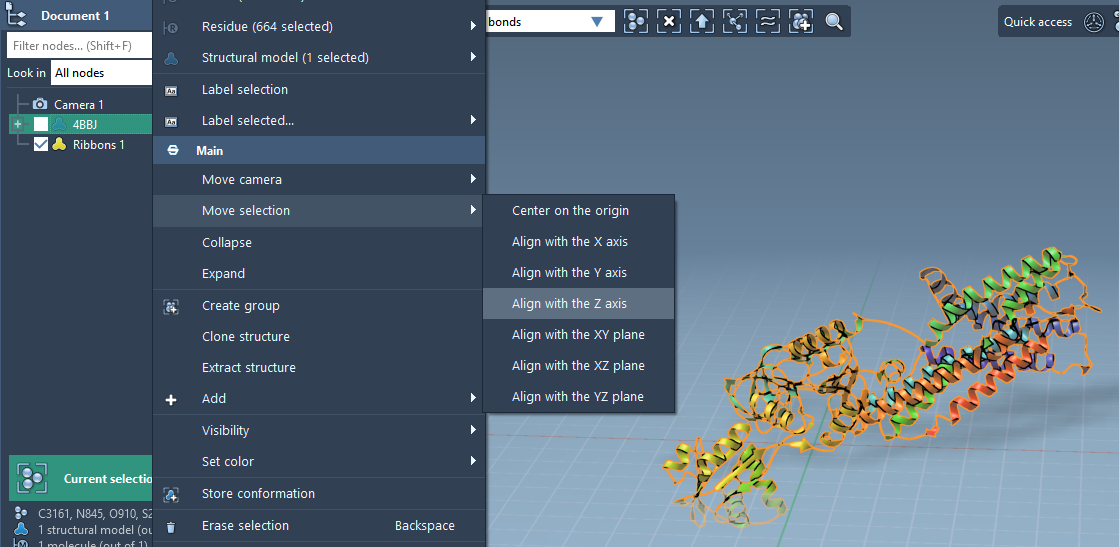
Aligning the Protein for Membrane Embedding
To build a membrane that makes sense biologically, it helps to align the protein’s transmembrane domain with the Z-axis. Here’s how to do this in SAMSON:
- Right-click on your protein in the Document view.
- Select Move selection > Align with Z axis.
- Then choose Move selection > Center on the origin.
Setting the Lipid Molecule
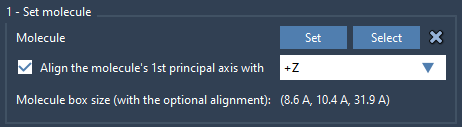
After aligning the protein, import a lipid molecule of your choice, such as POPC or DPPC. With the lipid selected in the Viewport or the Document view:
- Click Set in the Molecular Box Builder.
- Align the lipid’s principal axis with
+Z, so that all lipids will face the same direction relative to the protein.
Defining the Box to Fit Around the Protein
Now define the molecular box that will contain your lipid layer:
- Center the box around the protein molecule.
- Adjust
XandYdimensions to extend slightly beyond the protein footprint. - Set a small value for the margin to control lipid-lipid spacing accurately.
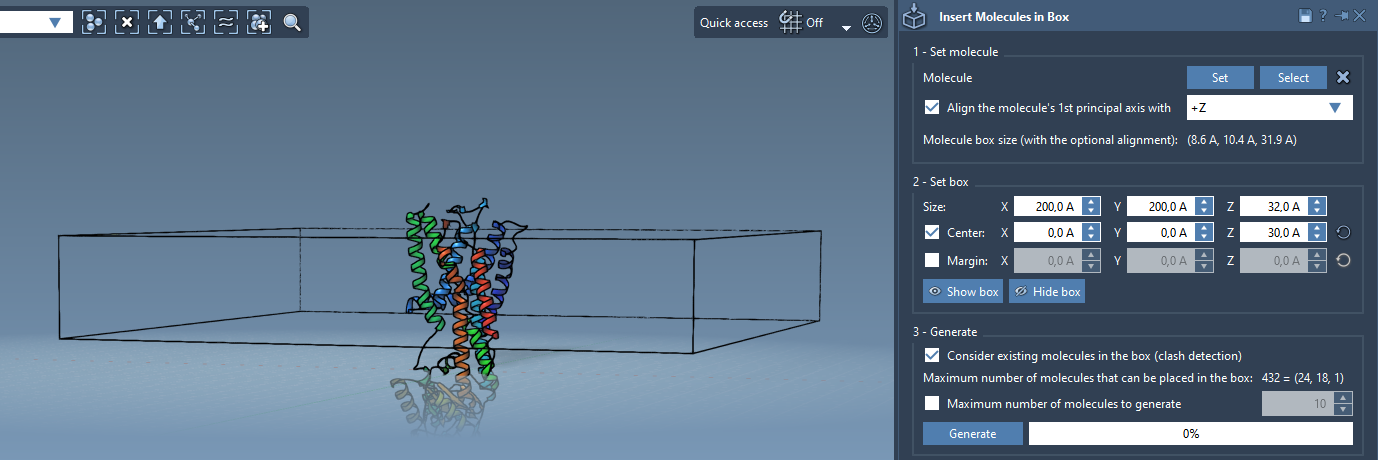
Generating the Lipid Layer
Before placing lipids, make sure to enable Consider existing molecules in the box. This ensures that lipids will surround the protein without overlapping it.
- Once settings are confirmed, hit Generate.
You will get a layer of lipids correctly placed around the protein, ready for further processing or simulation setup.

Going One Step Further: Bilayers
For those modeling bilayers, repeat the process:
- Generate the first layer aligned to
+Z. - Shift the box center downward.
- Generate a second layer aligned to
-Z.
Now you’ve got a full lipid bilayer built directly around your protein, ready for solvation, ion placement, or MD parameterization.
If you’re trying to streamline membrane system setup and want a GUI-first approach, Molecular Box Builder may save you hours of effort.
To learn more, visit the Molecular Box Builder documentation page.
Note: SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





