Building realistic molecular systems, such as protein–membrane complexes, is often a time-consuming and meticulous task in molecular modeling workflows. While several tools and techniques exist for assembling lipid bilayers, many require manual file editing or scripting, and often lack integration into a visual modeling environment.
If you’ve ever wanted to create a lipid bilayer around a membrane protein, without having to switch between multiple tools or guess at box dimensions, SAMSON’s Molecular Box Builder provides a straightforward, visual approach—right in your modeling workspace.
From Structure to Bilayer in Minutes
Let’s walk through how Molecular Box Builder helps you generate a single or double lipid layer around a membrane protein in just a few steps.
1. Orient Your Protein
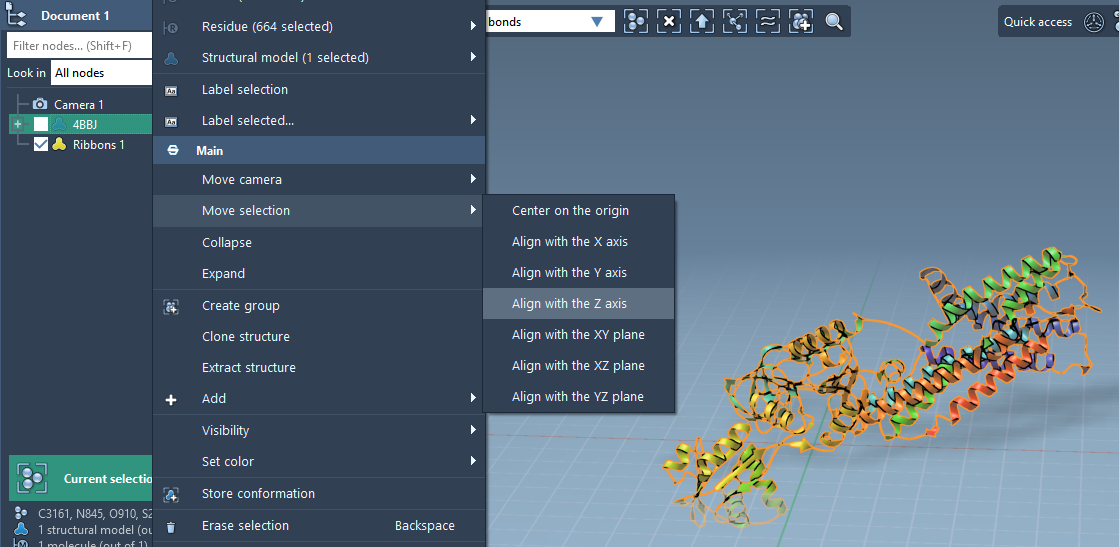
To begin, load your target protein and align it with the Z axis. In SAMSON:
- Right-click the protein in the Document view.
- Select Move selection > Align with Z axis.
- Then select Move selection > Center on the origin.
2. Introduce a Lipid Molecule
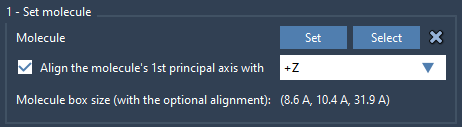
Next, load the lipid molecule you’d like to use for the bilayer (e.g., POPC). Select it in either the viewport or the document and click Set in the Molecular Box Builder.
To ensure proper orientation:
- Select
+Zas the alignment axis.
3. Define the Box
Set up a box that surrounds the protein:
- Enable Center and place the box around the protein.
- Adjust the height in the
Zdirection to match a single lipid layer. - Optionally, set a margin between molecules to fine-tune packing.
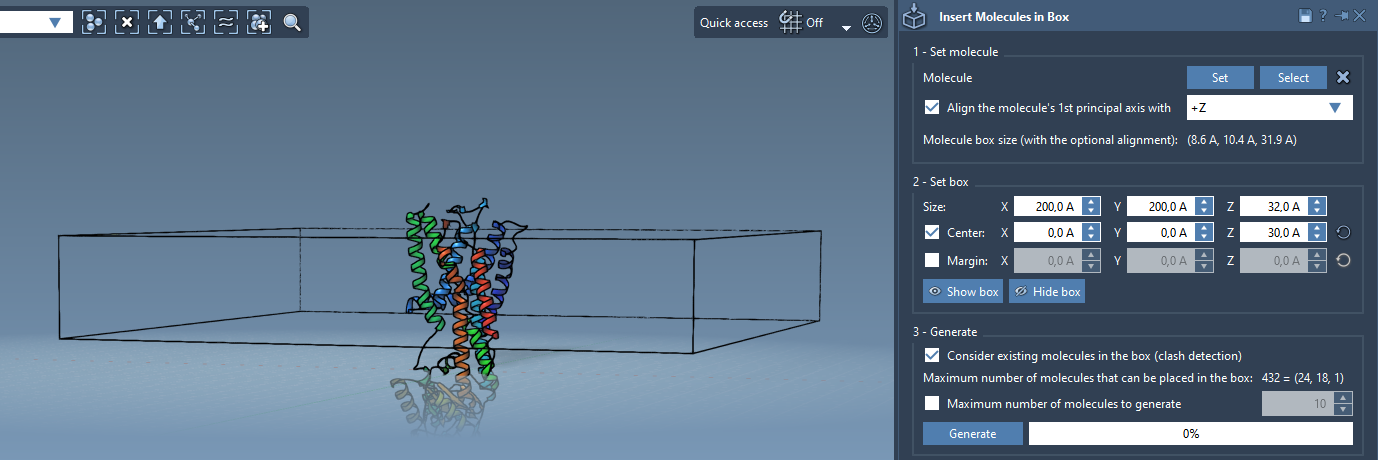
4. Generate One or Two Layers
Now comes generation:
- Check Consider existing molecules in the box to avoid placing lipids inside the protein.
- Click Generate.
You now have a single lipid layer around the protein:

To add a second layer:
- Shift the box along the
Z-axis using the center parameter. - Switch lipid alignment to
-Z. - Run generation again to complete the bilayer.
Intuitive, Reversible and Customizable
What makes this workflow effective is how seamlessly it integrates visual alignment, box definition, and generation. You can see exactly how your molecules are arranged and easily update dimensions or alignment without leaving the modeling environment. No scripts required. No trial-and-error packing.
If you’re preparing a protein–membrane system for simulations or structural analysis, this tool helps you focus more on science and less on setup.
To learn more, including how to prepare your system for simulation with GROMACS, check out the full documentation here:
https://documentation.samson-connect.net/tutorials/molecular-box-builder/molecular-box-builder/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





