When modeling molecular processes, capturing transitions between different conformations of a protein is often essential. Whether you’re exploring activation pathways, refining docking poses, or setting up umbrella sampling simulations, intermediate structures along these conformational paths are a valuable resource. But generating those realistic transitions can be a time-consuming task—especially when you’re doing it manually or relying on computationally expensive simulations.
This is where the ARAP Interpolator extension in SAMSON offers a useful alternative. ARAP stands for As-Rigid-As-Possible, a geometry-based approach to interpolate between two molecular conformations while maintaining structural realism. It’s fast, easy to use, and particularly helpful for creating starting points for simulations or for visualizing transitions.
Why Use ARAP Interpolation?
This method allows you to:
- Compute a smooth conformational path in seconds
- Retain biological plausibility through geometry-aware interpolation
- Export intermediate frames as PDBs for downstream analysis or simulation
- Use realistic reaction coordinates as input to molecular dynamics techniques
A Practical Example: From Closed to Open States
Let’s look at a real-world example used in SAMSON: computing the motion between closed and open conformations of the Diphtheria Toxin. The approach begins with downloading the two structures (1DDT and 1MDT), cleaning them, and defining their respective conformations. These become the start and goal inputs for interpolation.
From there, the ARAP app takes over. It matches atoms (usually all heavy atoms), and uses configurable strategies to build ARAP edges—essentially the framework for deformation. Options include bond-based, α-carbon adjacency, distance cutoffs, and more.
After defining the interpolation parameters (e.g., 20 intermediate steps) and initiating the run, a path is computed connecting the two conformations. Each step is accessible as a conformation object and can be exported or used in follow-up processing such as P-NEB, steered MD, or umbrella sampling.
Interpreting and Using the Output
The ARAP interface allows users to:
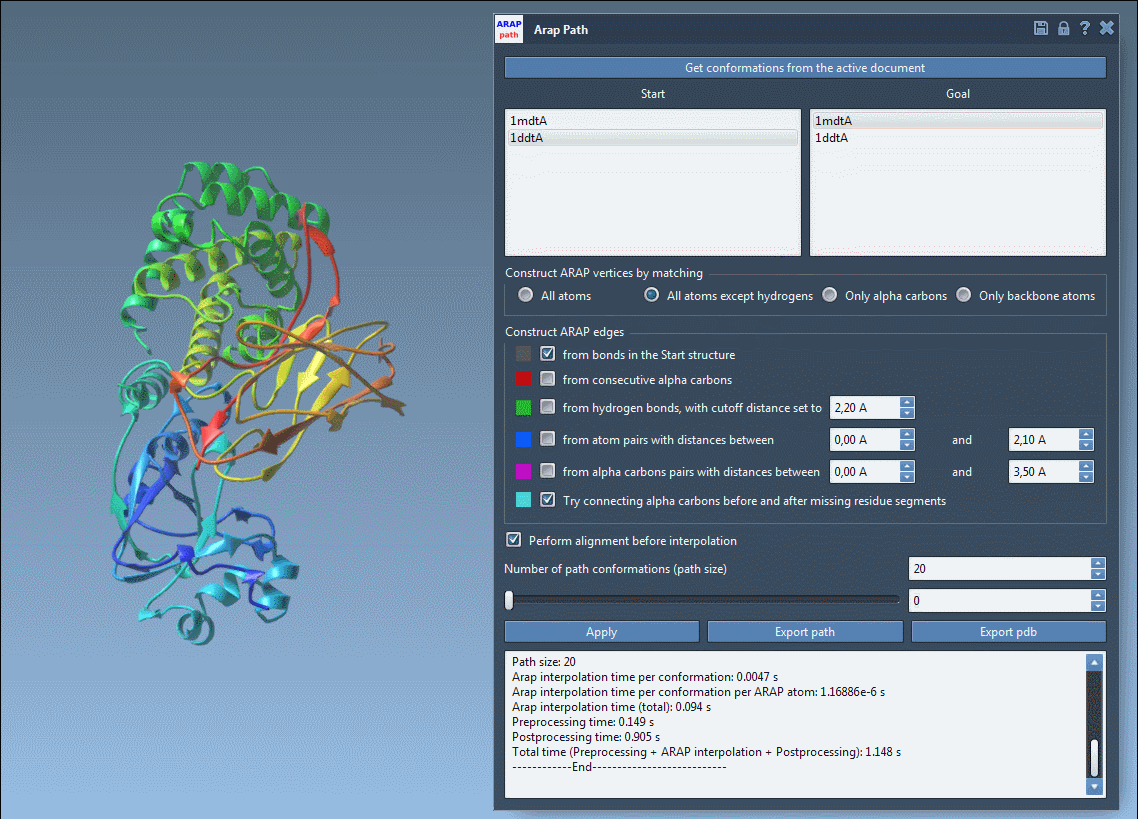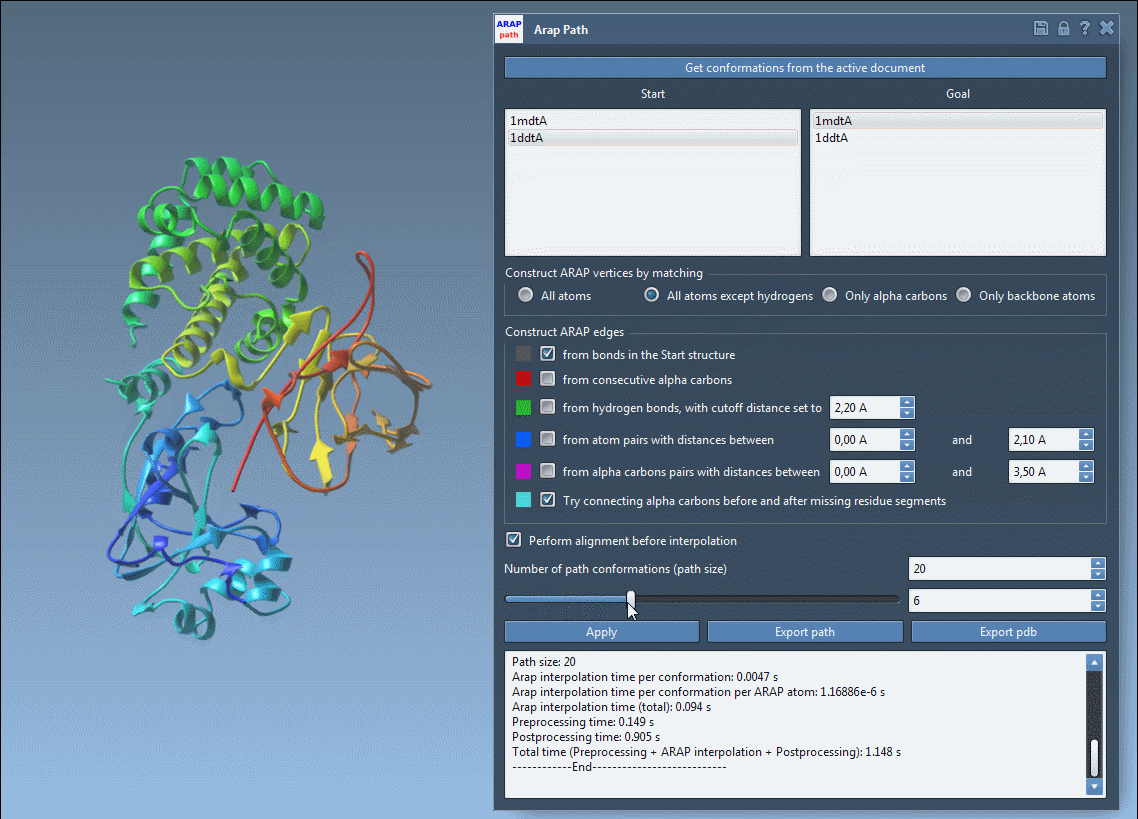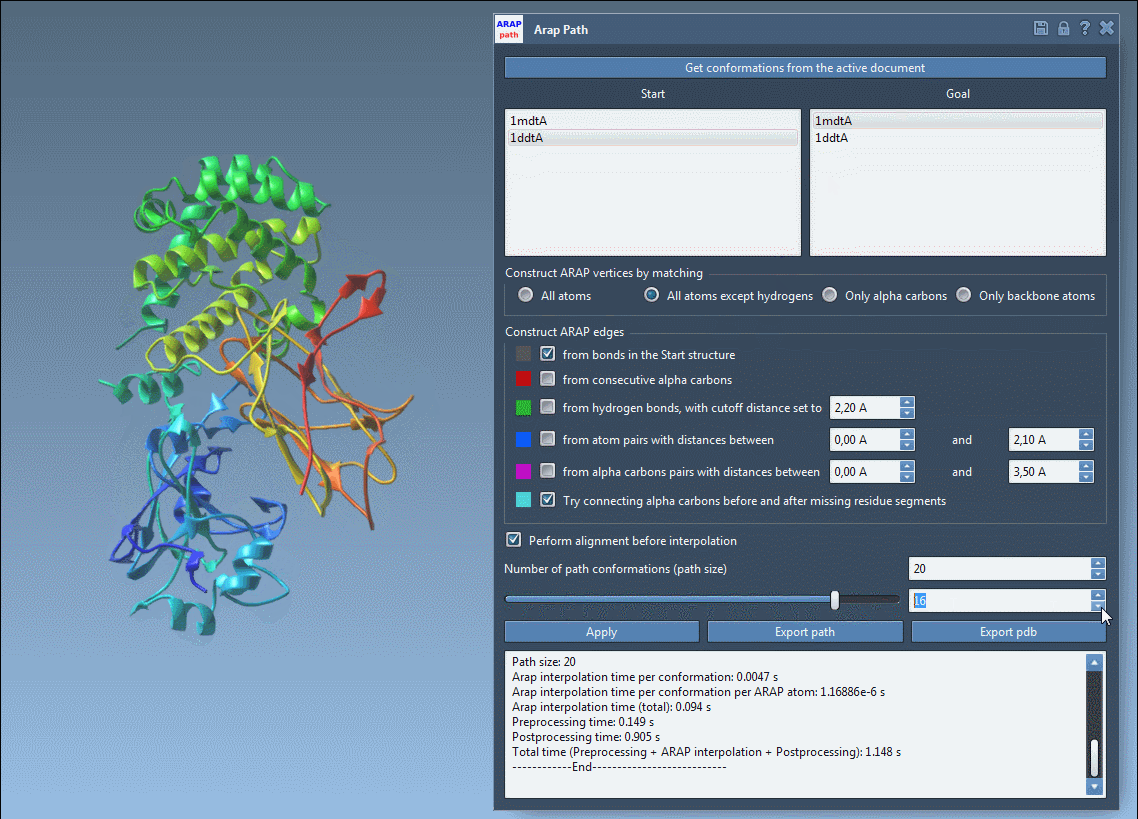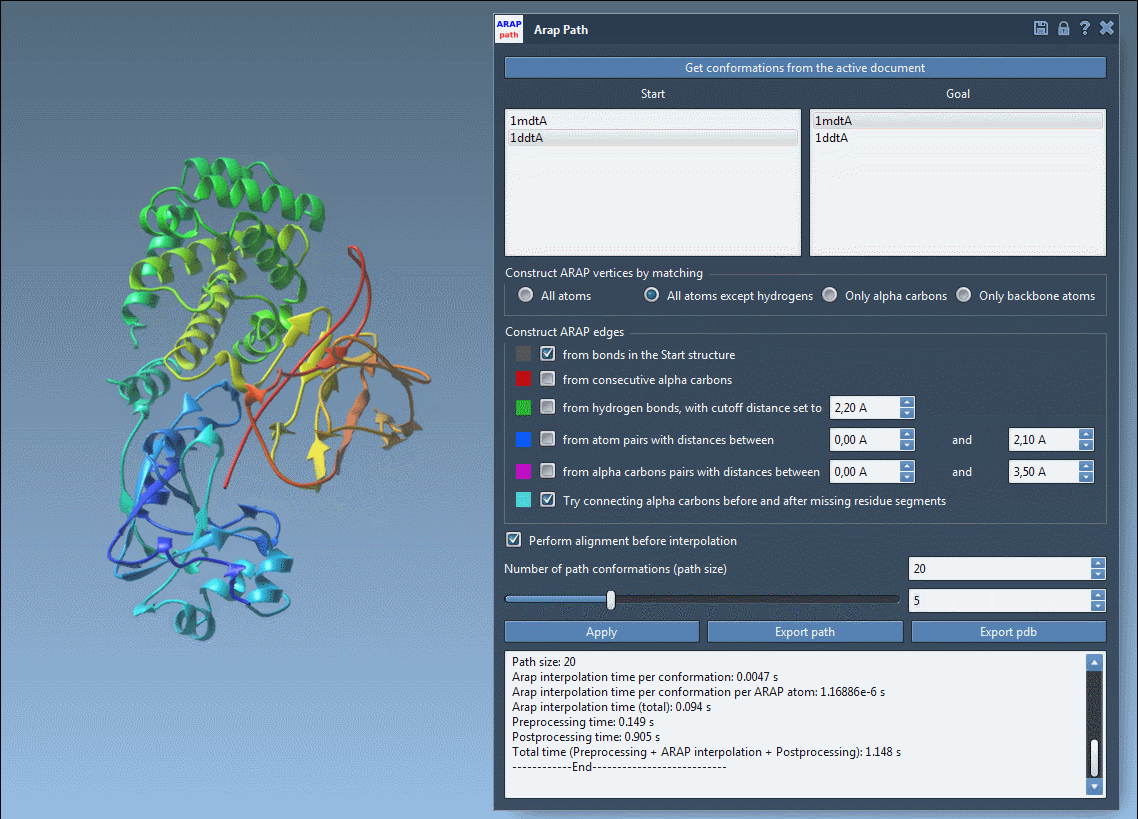
- Visualize interpolated conformations using a slider
- Toggle ARAP edges for a structural overview of how deformation propagates
- Export the path to PDB format—either as one file or separate PDBs per frame
This animated view of the transition provides a direct sense of the deformation process. For further analysis or simulation setup, the output structures can be used to define reaction coordinates or to extract frames for enhanced sampling strategies.
Things to Watch Out For
One common error occurs when molecular structures are not pre-processed properly. If you see the error: “Cannot proceed because the structure does not make one connected component”, it usually means that water molecules, ligands, or ions are still present. Be sure to use Home > Prepare in SAMSON to clean structures before interpolation.
Final Thoughts
If you’re spending too much time generating transition pathways manually—or waiting hours for full MD trajectories just to get intermediate conformations—this interpolation approach can save a lot of time. It’s fast enough to try ideas quickly and flexible enough to support advanced simulations later on.
To learn more and try it yourself, visit the official documentation: ARAP Interpolation for Protein Structures.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at www.samson-connect.net.





