When studying protein dynamics, researchers often encounter an important question: how do we realistically interpolate between two known conformations of a protein structure? Whether you’re setting up umbrella sampling simulations or exploring conformational transitions for molecular docking, creating a smooth and biologically meaningful path between two states can be challenging and time-consuming.
This blog post guides you through a quick and effective approach to generate such transitions using the As-Rigid-As-Possible (ARAP) Interpolator in SAMSON. You’ll discover how to move from loading structures, defining conformations, and launching interpolation to finally visualizing and exporting your results—all within a few minutes.
Why Use ARAP Interpolation?
- It computes continuous protein transitions in seconds.
- It helps build meaningful reaction coordinates for free energy simulations.
- It aligns and interpolates conformations using geometrically consistent methods.
Step-by-Step Guide: From Conformations to Interpolated Paths
1. Create Clean Conformations
After fetching your protein structures (e.g. 1DDT and 1MDT), make sure to clean them using Home > Prepare to remove ligands, ions, water, and alternate locations. This ensures the structures form one connected component, a requirement for ARAP interpolation.
Then, for each cleaned structure, create a named conformation like 1DDT A and 1MDT A using Edit > Conformation.
2. Launch the ARAP Interpolation App
Open the app through Home > Apps > Biology > ARAP Path Interpolation. Here, you’ll select your Start and Goal conformations.
3. Customize Matching and Connectivity Settings
- Matching Atoms: Choose All except hydrogens to restrict interpolation to heavy atoms.
- Edge Construction: Check boxes to create edges from bonds in the start structure and to bridge missing α-carbon segments.
- Alignment: Enable Perform alignment before interpolation to align conformations prior to interpolation.
- Path Length: Set Number of path conformations to 20 (or adjust depending on your needs).
Once configured, click Run. The app generates a transition path within seconds.
4. Analyze and Export the Path
The visual interface provides a slider to scrub through the interpolated conformations. You can examine how fragments move between frames and make use of visual edges to understand how connectivity drives motion.
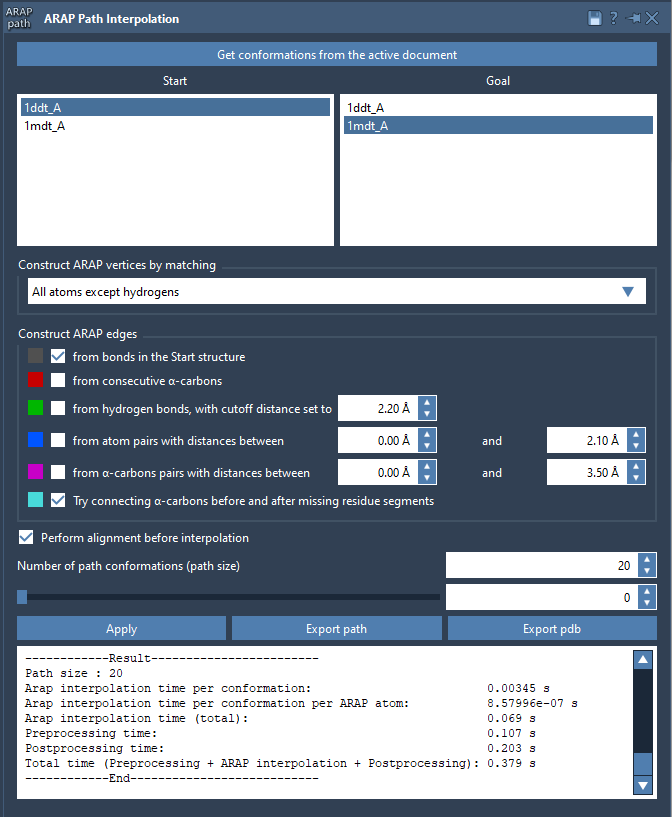
Edge types are color-coded, based on how the algorithm built connections. These can be toggled for visibility in the Document view.
5. Export Your Work
- Export path: Saves the full path as a trajectory within SAMSON.
- Export PDB: Outputs the entire path or each frame as a separate PDB file.
When to Use It
This interpolation proves useful when:
- Setting up umbrella sampling with tools like the GROMACS Wizard
- Preparing input paths for steered MD or P-NEB (Nudged Elastic Band) refinements
- Analyzing structural ensembles or performing dimensionality reductions
To explore detailed instructions and advanced options, visit the official documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. Try it out by downloading SAMSON at samson-connect.net.





