Designing polymers can be a repetitive task — literally. If you’ve ever needed to build a long polymer with a specific repeating pattern, manually connecting monomers one by one can quickly become frustrating and error-prone. Whether you’re modeling materials or biological macromolecules, repeating units show up frequently. That’s exactly where SAMSON’s Polymer Builder comes in, making the process of registering and reusing customized monomer sequences extremely practical.
In this post, we’ll explore how you can register monomer sequences that you can reuse throughout your polymer design workflow. It’s a small feature, but one that can save a lot of time — especially as projects grow larger and more complex.
What’s a Monomer Sequence?
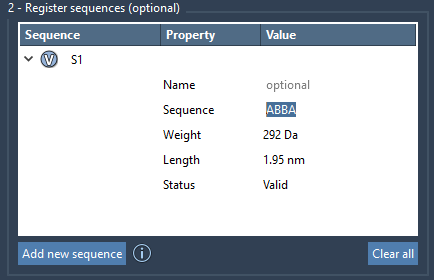
Once you’ve registered individual monomers in SAMSON, you can group these into sequences — predefined patterns of monomer units that make up parts or all of your polymers. For example, if you have three monomers A, B, and C, you could define a sequence like ABBA and re-use that block throughout a polymer.
Registering a Sequence
Registering a monomer sequence is simple:
- Click Add new sequence in the Polymer Builder interface.
- Enter a sequence using monomer IDs, such as
ABBAorA=BBif you want a double bond between certain units.
This step creates a reusable block identified as S1, S2, etc. Monomers used will be highlighted when you click the V button next to each sequence.
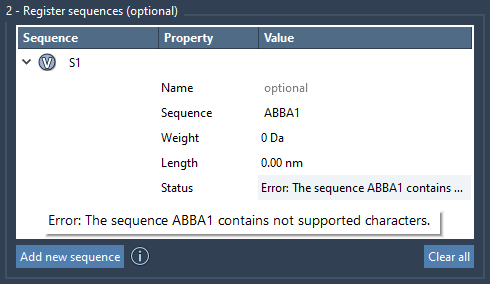
If you make a typo or reference a monomer that doesn’t exist, SAMSON will flag it clearly with an error in the Status column, helping you validate the sequence before using it.
Tuning Bond Types
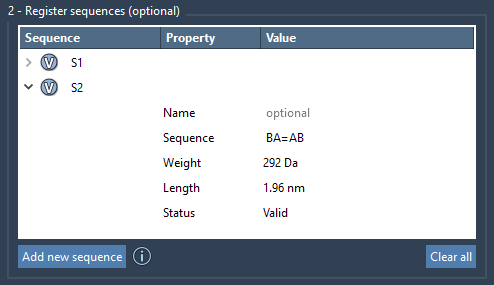
Sequences aren’t limited to just ordering monomers — you can also define the bond types connecting them. By default, bonds are single, but special syntax lets you modify the type:
A=B➝ A and B are connected by a double bond.B#C➝ B and C are connected by a triple bond.
This allows you to encode not only structure but chemistry directly into your design patterns without manual adjustments later.
Modifying and Reusing Sequences
Once registered, sequences can be edited directly within the Polymer Builder table. You can rename them, update sequences, change names for groupings, and see an immediate update in the predicted molecular weight and length of each sequence.
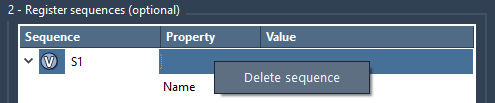
If a project shifts direction or a sequence is no longer needed, you can simply right-click on it and choose Delete sequence. To delete all, just click Clear all.
Why It Matters
Defining and using sequences isn’t just about saving time—it’s about enabling repeatability and consistency throughout molecular modeling workflows. For researchers modeling large biopolymers or materials scientists working with complex copolymers, sequences help establish modular building blocks, which can be tested, reused, and shared across projects.
This built-in feature also aligns well with simulation tools like GROMACS and LAMMPS, since reproducibility and structural logic are essential for accurate modeling and data generation.
Explore more about polymer workflows and advanced functionality in the full Polymer Builder documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





