One of the most common questions molecular modelers have when setting up docking experiments is: “Where should I define the search space?” Whether you’re targeting a known binding pocket or screening broadly across the protein, defining the search domain correctly is essential for efficient and meaningful docking results.
In SAMSON, thanks to the AutoDock Vina Extended extension, you have flexible and visual control over the definition of your search domain. Here’s a concise guide to help you streamline this critical step.
Why the Search Domain Matters
The search domain in a docking setup represents the 3D box where the software will explore possible ligand conformations and positions. If it’s too small, you may miss the right pose. If it’s too large, computations take longer and may yield noisy results.
How SAMSON Helps
SAMSON provides dynamic options for setting the search box in your docking experiments. You can:
- Adjust the box manually in the 3D Viewport by dragging its corners.
- Define it based on the receptor’s entire structure (default).
- Set it based on a selection or known binding site, for more localized exploration.
Step-by-Step: Setting the Search Domain Based on a Known Binding Site
- First, load your system into SAMSON. Ideally, this includes the target protein and a known bound ligand.
- Select the known bound ligand in the Document View.
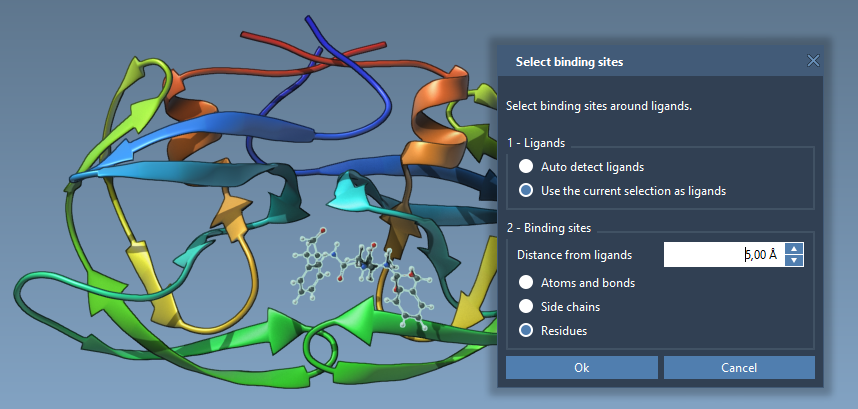
- Click Select > Biology > Binding sites. Choose your parameters (e.g. atoms within 5 Å radius) and confirm. This selects surrounding residues.
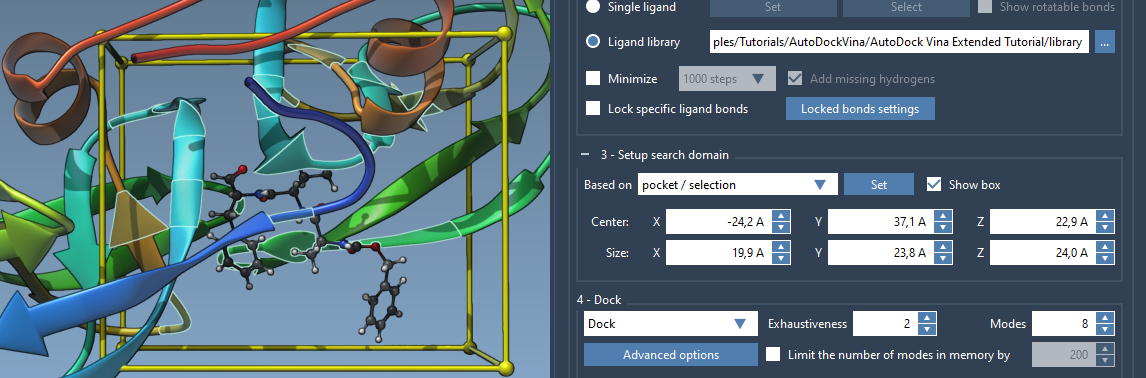
- From the AutoDock Vina Extended app interface, under Search domain, choose Based on pocket/selection from the drop-down list and press Set.
Now, the yellow box in the Viewport will tightly surround your region of interest, ensuring focused docking around the expected binding site.
Prefer to tweak things manually? You can still drag the sphere handles on the box corners to resize or reposition the domain directly in the Viewport.
Tips for Better Docking
- Smaller, well-placed search domains result in faster computations and often better scoring results.
- For unknown binding sites, start broad, then refine once promising interactions are found.
- Make use of ligands already co-crystallized with your protein – they are great guides for site selection.
Tip
If you don’t see the search domain box in your Viewport, check or enable the Show box option in the AutoDock Vina Extended interface.
Visual Feedback Metters
The live visualization in SAMSON makes it easy to confirm that your docking search box covers the right region. This is especially handy for large proteins with multiple potential sites.
Want to explore more about running docking workflows, analyzing interactions, and preparing receptors? Visit the original tutorial: AutoDock Vina Extended tutorial.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





