Working with large molecular systems often means wading through overwhelming layers of atoms, bonds, residues, chains, and structural groups. For molecular modelers and structural biologists, one of the most common tasks is to isolate particular regions of interest – ligands, residues in helices, or atoms in a specific chain. Spending time manually clicking through the hierarchy in complex biomolecular structures can make analysis inefficient and error-prone.
Thankfully, SAMSON provides a streamlined solution through its Node Specification Language (NSL), which lets you filter and select specific nodes directly in the Document View. This post focuses on using NSL for filtering nodes in the Document View – a straightforward yet powerful feature that can make your workflow more efficient.
Why Filter with NSL?
The Document View in SAMSON displays all the nodes (atoms, chains, residues, etc.) as a hierarchy. However, in complex structures, this can become unwieldy. Filtering nodes allows you to instantly narrow down what you see and interact only with the relevant parts of your system. For example, you may want to:
- View only ligands or side chains
- Isolate all residues of a certain type (e.g., all alanines except one)
- Identify atoms beyond a certain distance from a selection
- Find structural groups having a particular atom type (e.g., sulfurs in side chains)
How Does It Work?
In the Document View, a filter field accepts NSL expressions. These expressions let you specify the type, attributes, and spatial relationships of nodes. Here’s a simple example:
|
1 |
n.t sg |
This expression filters node.type structuralGroup, making it easier to review only structural groups, as shown below:
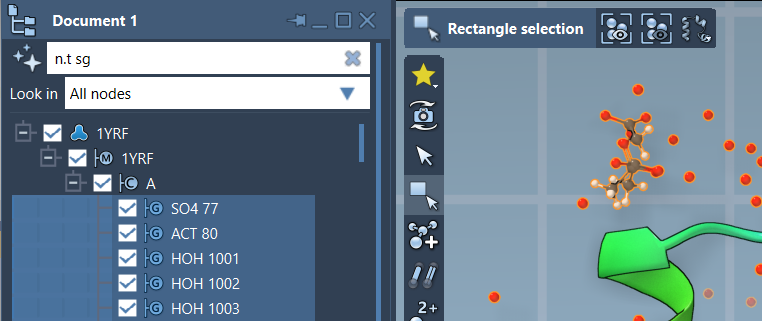
Once filtered, you can hit Enter to select the matching nodes directly from the list.
Examples of Useful Filters
n.t r and not r.t ALA: all residues except alaninen.t sc h S: side chains that have sulfur atomsr.ss h: all residues in alpha helicesC w 5A of "GLN 2": carbon atoms within 5 angstroms of residue GLN 2
Smart Assistance
Not comfortable writing filter expressions? Click the ⭐️ Ask AI button next to the filter field. The SAMSON AI Assistant understands your document’s hierarchy and can help generate NSL expressions tailored to your molecular model.
From Filter to Selection
Filtered nodes can be selected in bulk for further operations: visualization tweaks, measurements, or custom scripting. Instead of manually picking parts of your structure, filtering and selecting via NSL turns this into a two-second task.
Final Thoughts
If you often find yourself overwhelmed by large molecular documents, learning to filter nodes using NSL in SAMSON can save time and reduce guesswork. This feature becomes particularly useful in structural biology, drug discovery, and any setting involving complex molecular assemblies.
To learn more about NSL, its full syntax, and numerous real-world examples, visit the official documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





