For molecular modelers working with free energy calculations, computing the Potential of Mean Force (PMF) can be a challenging and often error-prone process. One of the most common bottlenecks is merging umbrella sampling results into a reliable PMF profile. Errors in folder organization, confusion around setting correct bounds and units, or difficulties in ensuring consistent reaction coordinates can often impede progress.
If you’ve ever found yourself manually editing WHAM inputs, double-checking filenames, or re-running simulations due to incorrect histograms — this post might save you some time.
The GROMACS Wizard inside SAMSON includes a dedicated tab for WHAM Analysis. This significantly streamlines PMF computation from umbrella sampling outputs, making it easier to perform and interpret PMF computations correctly.
Getting Started with WHAM Analysis
After completing your umbrella sampling simulations — either manually or using the automated batch computation feature — switch to the WHAM Analysis tab in the GROMACS Wizard.
You can select your project path manually or use the convenient auto-fill option ![]() to reuse the setup from prior steps. The folder structure should contain subfolders (typically numbered) and simulation results for various umbrella windows.
to reuse the setup from prior steps. The folder structure should contain subfolders (typically numbered) and simulation results for various umbrella windows.
Setting Parameters, Visualizing Results
The Wizard will extract key information from your umbrella sampling results, including reaction coordinate names, time and temperature parameters.
You can then:
- Choose a reaction coordinate
- Adjust time bounds or energy units
- Modify custom bounds if needed
Click Compute to run the WHAM algorithm. The Wizard will generate two useful visualizations:
- PMF plot
- Histogram of sampled windows
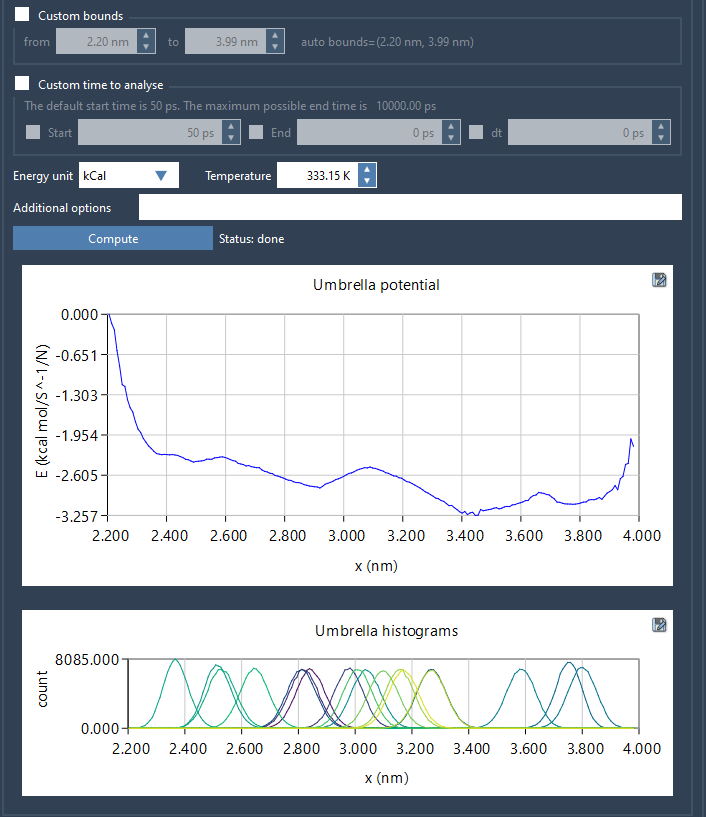
The PMF visualization allows you to inspect the energy profile along your reaction coordinate. Importantly, the histogram displays how well-sampled each region is — if you see major gaps or uneven distribution, it could be a sign that additional umbrella windows are needed.
Time-saving Reuse of Results
The GROMACS Wizard automatically saves results in a wham_results subfolder. If you switch reaction coordinates, the Wizard reuses previously computed data when available — so you don’t end up recalculating unnecessarily.
Why It Matters
If you’ve used WHAM via the command-line before, you know how easy it is to mistype inputs, mislabel files, or spend hours debugging. By organizing simulation data visually and automating key steps, this tool helps you stay focused on interpreting results rather than wrangling text files.
To learn more or explore the complete tutorial, visit the original documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON from https://www.samson-connect.net.





