When preparing protein structures for simulation or modeling, one frequent challenge is identifying residues that adopt energetically unfavorable conformations. Even a few outliers in backbone dihedral angles can introduce strain, reduce model quality, or degrade simulation stability. That’s where the Interactive Ramachandran Plot in SAMSON can make a difference.
Instead of manually checking dihedral angles or exporting data to external plotting tools, you can now explore, validate, and even edit protein conformations directly within an intuitive visual interface — with live feedback on the 3D structure.
What the Interactive Ramachandran Plot Offers
A Ramachandran plot is a 2D graph that maps the φ (phi) and ψ (psi) backbone dihedral angles of amino acid residues in a protein structure. It highlights which angle combinations are energetically favorable (typically yellow zones), borderline, or disallowed (white zones).
But this isn’t just a static plot. In SAMSON, each point on the plot is clickable — and editable — giving you a fast way to spot problem areas and refine them visually.
Step-by-Step: Validating Residues in One Click
Once you’re in SAMSON and you’ve installed the Interactive Ramachandran Plot Extension:
- Load a protein (e.g. PDB ID
1YRF) via the Home > Fetch panel. - Launch the plot app from Home > Apps > Biology > Ramachandran Plot.
- Click Update to generate the plot.
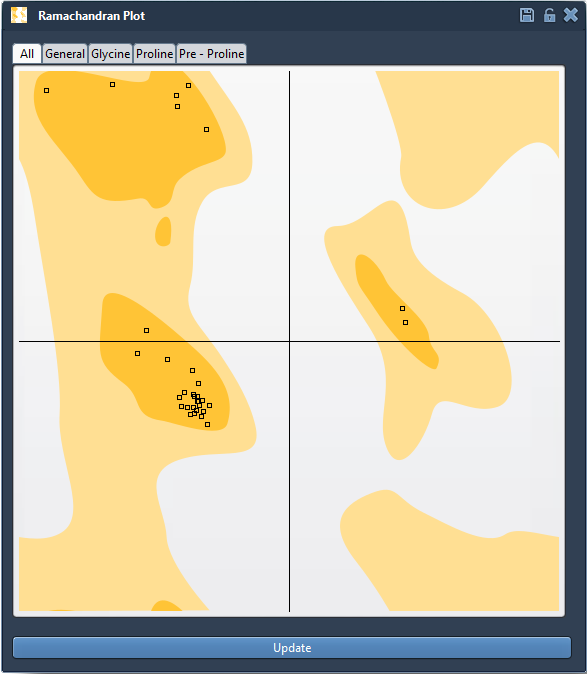
Residues are categorized into tabs (General, Glycine, Proline, Pre-proline). This categorization helps you focus on specific residue types prone to unusual conformations.
Explore and Fix Outliers Faster
Click any dot on the plot to immediately select the corresponding residue in the 3D viewport. The dihedral angles appear in the status bar, so you can inspect the actual φ and ψ values.


Want to Adjust a Residue? Do It Visually
If you find a residue in an unfavorable region, you can make real-time adjustments:
- Option 1: Drag the point in the plot itself. The 3D protein structure will update automatically.
- Option 2: Use the Twister editor from the left-hand viewport toolbar to tweak the residue directly in 3D space. The plot updates simultaneously.
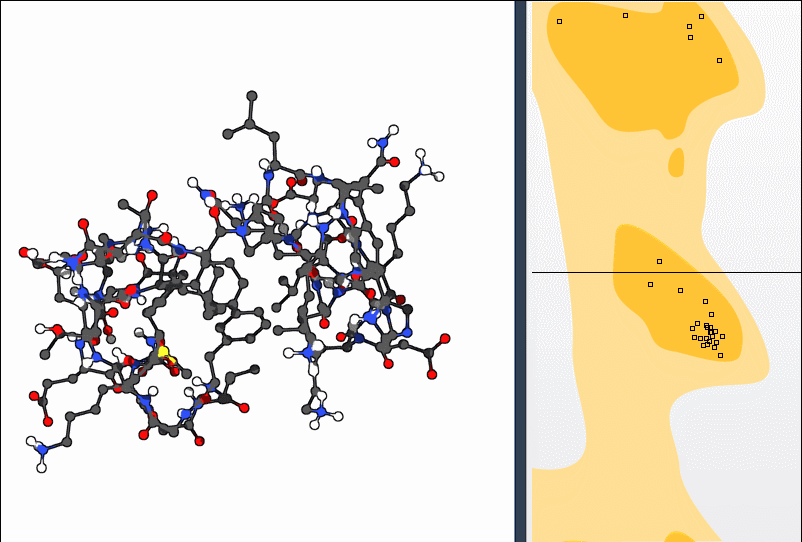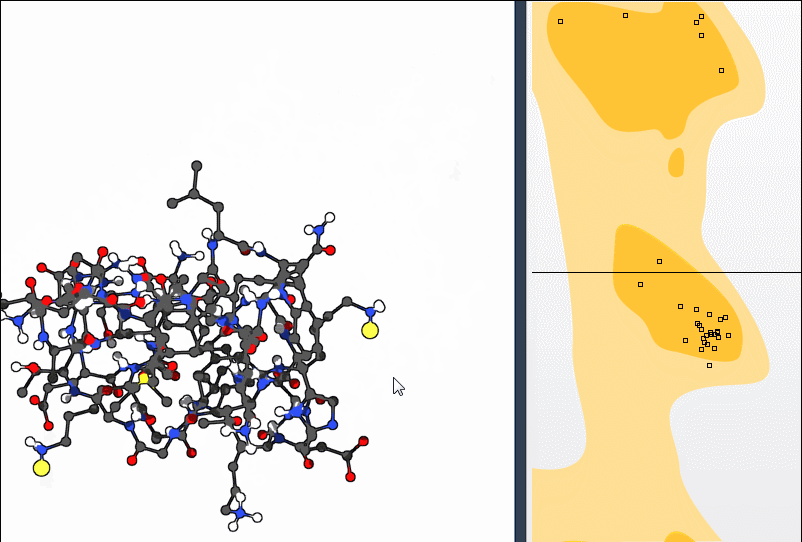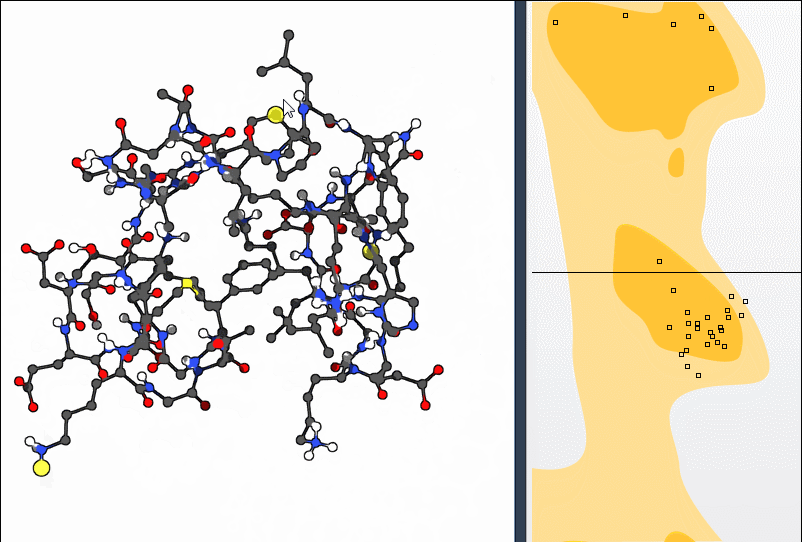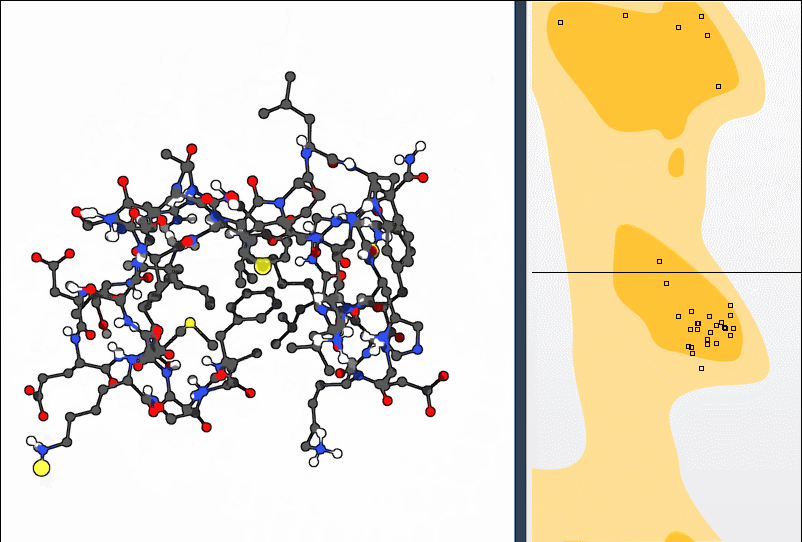
Need to undo a move? Just press Ctrl/Cmd + Z.
Why It Matters
Misplaced residues can lead to:
- Simulation artifacts and slow convergence
- Incorrect recognition by active sites or docking agents
- False-positive results when predicting flexibility
By interactively validating residue placement, you reduce pre-simulation headaches and improve the reliability of your results.
To learn more, visit the full documentation page: Interactive Ramachandran Plot – SAMSON Documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





