Understanding molecular interactions often requires not just static snapshots of a system, but watching how molecules move—especially when investigating mechanisms like ligand unbinding or conformational transitions. One common challenge for computational chemists and molecular modelers is the need to export only a subset of atomic coordinates along a precomputed path. Whether for plotting a reaction coordinate, prepping input for enhanced sampling techniques, or compiling frames for visualization tools, selectively exporting atoms along a trajectory can save time and avoid post-processing headaches.
The Export Along Paths extension in SAMSON offers a simple workflow to export just the atoms you need—for example, a ligand—along predefined paths. Here is how it works, using a sample system of Lactose permease and its ligand TDG, already equipped with unbinding paths computed using the Ligand Path Finder.
Step-by-Step: Exporting Atom Trajectories for a Subset
Start by loading the sample system:
- Go to Home > Download in SAMSON.
- Paste this document link: https://www.samson-connect.net/documents/046f1acd-c799-40f6-8185-cb4847eff795
- Click Download and open the document.

Once the document is open, follow these steps to export a subset of atoms:
- Open the Export Along Paths App from Home > Apps > All > Export Along Paths or using Shift + E.
- Click the Advanced panel to expand subset options.
- In the Document view, select the ligand (
TDG).

- Click Add to register this as a model for export.
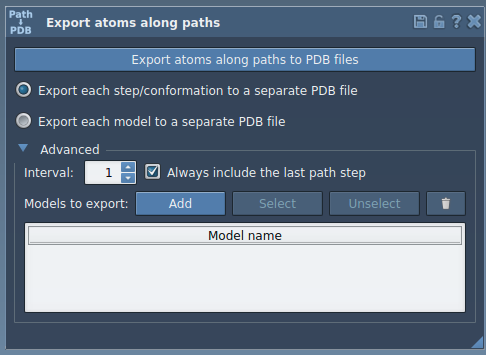
This model will appear in a table. You can:
- Rename it by double-clicking the name.
- Add multiple models (e.g., protein backbone, water molecules).
- Reset or deselect models using the contextual menu.

When you’re ready:
- Select path(s) you want to use.
- Choose export format: all frames in one PDB, or multiple PDBs (one per frame).
- Click Export atoms along paths to PDB files.
SAMSON will prompt you to choose a destination folder and file prefix.
Why This Helps
By exporting only the atoms you’re interested in, you avoid bloated files and reduce post-export filtering. This approach is especially helpful for generating refined datasets for reaction coordinate analysis, visualization of ligand paths, or preparing input for methods such as umbrella sampling or metadynamics.
This method is also useful when multiple atoms—like a binding site region or backbone atoms—need to be tracked together. Multiple models can be exported in a single session, offering flexibility for complex workflows.
To learn more about Export Along Paths and access complete step-by-step guidance, visit the dedicated documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at www.samson-connect.net.





