Tracking molecular motion can be essential in areas ranging from free energy calculations to visualizing ligand unbinding. Yet, if you’ve ever tried to get a clean set of coordinates for just a ligand traveling through a protein tunnel, you know how overwhelming it is to sift through thousands of atoms in every trajectory snapshot. SAMSON makes this much easier—particularly with the Export Along Paths extension.
Here’s how you can export just the atoms that matter—like a ligand moving along a path—so you can analyze or visualize precise movement during simulations without getting bogged down in excess data.
🎯 The Problem
Imagine you’ve computed a ligand pathway with something like the Ligand Path Finder. You’re ready to simulate or analyze the unbinding route. But your tools need input: a sequence of PDBs showing your ligand as it follows that path. Exporting all atoms in the system is often unnecessary and inefficient, especially when you’re only interested in a ligand or a small subset of atoms. That’s the problem the Export Along Paths extension solves very practically.
🔬 Exporting a Subset of Atoms Along a Path
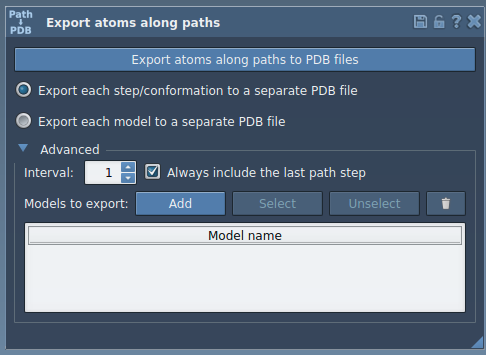
After installing and launching the Export Along Paths extension within SAMSON, here’s how you isolate and export only the atoms you care about—without writing a line of code.
- In the Document View, select your molecule of interest (for example, the ligand
TDGthat comes with the tutorial system). - Expand the Advanced section in the app interface.
- Click Add to define your selection as the model you wish to export. It shows up in a table below.

You can rename the model by double-clicking its name in the list, and you can define multiple models if needed.

Now that the model is defined, you can select your path(s) in the document, then choose whether you want the output as a single PDB (with multiple frames) or one PDB per frame. Click the export button, and that’s it—you’ll be prompted to choose a destination folder and a filename prefix.
🔍 When to Use This
This targeted export is useful anytime you’re more interested in an atom group’s motion rather than in the entire molecular system. Typical examples include:
- Ligand movement through a binding tunnel
- Molecular switches or specific side chain motion
- Inputs needed for reaction coordinate-based computations
- Preparing data for enhanced sampling methods
It helps you focus on the molecular components of interest, makes file sizes manageable, and streamlines integration into downstream analysis tools that require series of PDB formats.
To explore more options and variations, consult the full documentation page: Export atom trajectories along paths.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at www.samson-connect.net





