One of the most common setup pains in molecular modeling arises when preparing atomic coordinates along a ligand’s exit or binding path. Whether you’re running free energy calculations, benchmarking enhanced sampling methods, or analyzing transition mechanisms, a clean and flexible way to export atom trajectories is invaluable.
SAMSON’s Export Along Paths extension offers a solution specifically designed for this purpose. It allows you to export the positions of selected atoms—like ligands, active site residues, or key backbone atoms—as they follow a predefined path. This is particularly useful when working with ligand unbinding trajectories generated via the Ligand Path Finder, or any computed transition path.
Why export a subset, not the whole system?
Typical molecular systems are large, and exporting the entire system at every frame along the path is not only file-heavy but also unnecessary for most downstream analyses. What if all you want is the unbinding ligand’s coordinates?
Thanks to SAMSON’s interface, you can easily define which atoms to track and export as they move. Here’s how to do this using the advanced panel in the Export Along Paths extension.
Step-by-step: Exporting a subset
- Open the Export Along Paths app from Home > Apps > All or press Shift + E.
- In the advanced interface, select the atoms of interest in the Document view. For instance, you might select the ligand
TDG.

- Click Add to define this selection as a model to export.
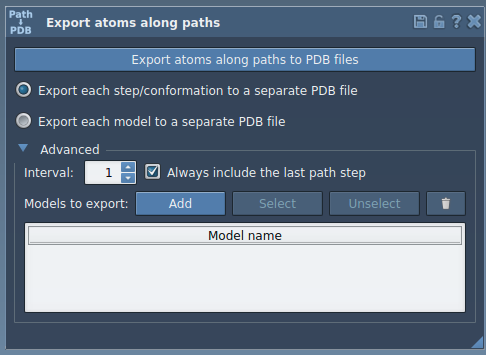
This adds the atom group to an export list. You can:
- Rename groups (e.g.,
Ligand_Unbinding_1) for better organization. - Add multiple atom groups, if needed.
- Select/deselect atom sets for export directly in the table.

Final Export
Once your models are defined and your path is selected:
- Choose your output format: a single PDB file containing all frames, or one PDB file per frame.
- Click Export atoms along paths to PDB files.
- Select your destination folder and name prefix.
This subset export is efficient and tailored for use cases where targeted information is preferred over full-system dumps—ideal for free energy profiles, ligand binding studies, and simple animations.
Use cases
- Create input for umbrella sampling and other enhanced sampling methods.
- Visualize only the ligand or region of interest along a transition path.
- Construct reduced representations for educational or presentation purposes.
Learn more in the full documentation page: Export atom trajectories along paths.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





