Free energy calculations of ligand binding or unbinding can be extremely powerful—but preparing input files often feels more painful than productive. One recurring challenge is extracting the atomic coordinates of molecules along a computed reaction path. If you’ve ever run enhanced sampling simulations or reaction coordinate studies, you know how cleaning up multiple trajectories and converting them into the right input format can be a bottleneck.
Thankfully, SAMSON includes a free extension called Export Along Paths that streamlines this process. In this post, we’ll walk you through how to extract the coordinates of a ligand moving along a path and save them into PDB files—perfect for free energy methods like umbrella sampling, WHAM, or other specialized calculations. 🧪
Why export paths for selected atoms?
Let’s say you’ve computed the extraction path for a ligand using the Ligand Path Finder extension. Perhaps you’ve optimized the trajectory with the parallel nudged elastic band approach. The next logical step is to export the ligand’s coordinates along that path as discrete frames. You might use these as windows for umbrella sampling, or to analyze conformational changes along the unbinding process.
Rather than saving full system trajectories—which can be bulky and hard to parse—you can focus just on the ligand. This reduces noise and gets you exactly what your workflow requires.
Step-by-step: Exporting only the ligand coordinates
1. In the Document view, select your ligand (in the tutorial case: TDG). Use selection tools as needed.

2. Open the Export Along Paths app via Home > Apps > All > Export Along Paths.
3. In the app interface, expand the Advanced section and click Add. This defines your selected atoms (i.e., the ligand) as the group to export.
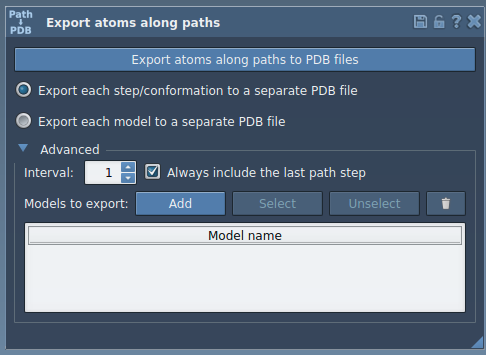
You’ll now see a table listing your defined export model. Rename it if necessary by double-clicking the name. You can also add multiple atom sets if needed.

4. Choose your preferred export format:
- Single PDB file with all frames
- Separate PDB files for each frame
5. Select the path(s) you want to export along in the Document view, then click Export atoms along paths to PDB files. You’ll be prompted to choose a destination folder and filename prefix.
That’s it. You now have clean ligand trajectory files ready to feed into whatever free energy protocol you follow. ✨
When should you use this?
This export procedure is ideal when:
- You’re preparing input for umbrella sampling or other enhanced sampling methods.
- You want to extract clean ligand-only trajectories.
- You’re scripting or automating post-processing and need lightweight, structured input formats.
To learn more, refer to the original documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. Get SAMSON at https://www.samson-connect.net.





