Molecular modelers frequently need to analyze how ligands move along defined pathways — whether for reaction coordinate studies, intermediate conformation extraction, or free energy calculations. But what if you’re only interested in the ligand, not the whole system? Exporting entire protein-ligand complexes frame by frame can be overkill, especially when your downstream workflow focuses on the ligand’s behavior.
The Export Along Paths extension in SAMSON offers a useful feature to export only a subset of atoms — such as a ligand — as it evolves along a computed path. Here’s how you can take advantage of this feature and streamline your molecular trajectory exports.
Getting Started
- Install the Export Along Paths extension.
- Restart SAMSON to activate the extension.
- Download the sample system for this tutorial (Lactose permease with the ligand TDG).
- Launch the Export Along Paths app via Home > Apps > All > Export Along Paths, or use Shift + E.
Selecting Just the Ligand
Once your sample system is open:
- Use the Document view to select the ligand
TDG—you can find it listed in the model hierarchy. - Expand the Advanced panel in the Export Along Paths app.
- Click Add to define the selected atoms as a model to export.
You’ll now see a table listing your custom model, which you can rename (e.g., “LigandTDG”) by double-clicking on its name.

You can define multiple models like this. For example, select and add the binding site residues if you want to track them separately.
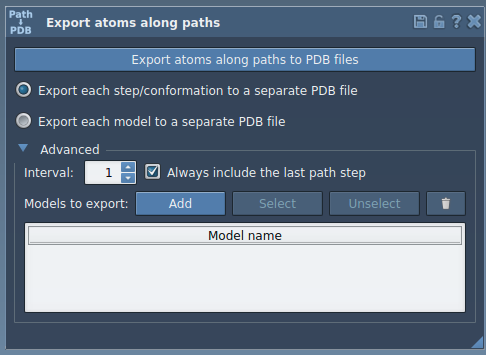
Setting Export Options
Before exporting:
- Choose your export format: either a single PDB with all frames or separate PDB files for each frame.
- Select the relevant path(s) along which the ligand moves.
- Set the frame interval in the Advanced section if you don’t need every frame.
Once set, click Export atoms along paths to PDB files. You’ll be prompted to choose a file destination and a name prefix — your ligand’s trajectory is now ready for GROMACS simulations, energy profiling, or visualization in your preferred tool.
Why This Matters
Exporting a focused subset of atoms along a path saves time, file space, and effort. It helps modelers stay focused on the molecular subunits they care about, keeping simulations light and relevant — especially in free energy workflows or when prepping reaction coordinates for analysis.
To learn more, including how to export all atoms or view paths, visit the original documentation: Export atom trajectories along paths in SAMSON.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON here.





