When modeling biomolecular systems, it’s common to compute pathways for ligand binding or unbinding. Whether you’re performing free energy profiling, setting up enhanced sampling simulations, or simply visualizing molecular movement, you often only need the atomic coordinates of a ligand—not the entire system—along a path. Exporting just what you need can save time, reduce data size, and simplify your analysis workflows.
The Export Along Paths extension in SAMSON lets you do exactly that. In this post, we walk you through how to export the 3D coordinates of a subset of atoms—such as a ligand—along a predefined path.
Why export only part of your system?
In complex molecular systems, exporting the entire atomic structure during a transition often leads to bulky files and unnecessary post-processing. But if you’re aiming to:
- Prepare a reaction coordinate file for free energy calculations,
- Track only a ligand moving along an unbinding path,
- Feed the coordinates into custom analysis scripts,
then exporting just a model—a group of relevant atoms—makes a lot of sense.
Step-by-step: Exporting a Subset of Atoms
After you’ve loaded your system and computed a path (e.g., using the Ligand Path Finder), here’s how to export the ligand’s trajectory along that path.
1. Open the Export Along Paths App
You can find it under Home > Apps > All > Export Along Paths or use the quick access menu (Shift + E).
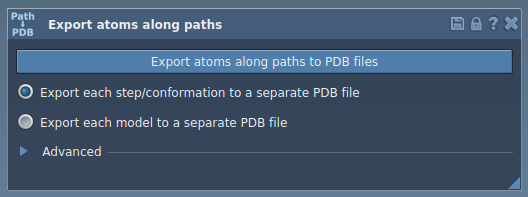
2. Select the Ligand
In the Document view, select the ligand (in our example, TDG) you wish to export.

3. Define Export Model
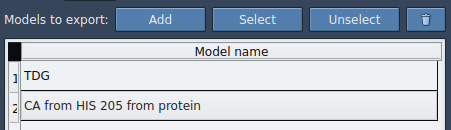
In the app, expand the Advanced panel and click Add. This creates a named model from your selection.
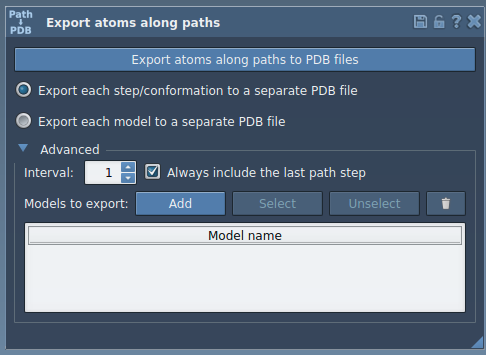
You can double-click the name to rename the model, or add other selections if needed.
4. Choose Export Options and Save
Select the path(s) of interest in the Document view, then choose whether to export each frame as a separate PDB file or a single file with all frames. Click Export atoms along paths to PDB files, set your destination—and you’re done ✅.
Benefits and Use Cases
- 🧪 Generate reaction coordinates for umbrella sampling or WHAM.
- 🎯 Focus on ligand-only dynamics without full system noise.
- 📂 Create compact datasets suitable for scripting or archiving.
This focused export functionality is also helpful for visualizations or when sharing minimal datasets with collaborators (or reviewers!).
To learn more, visit the full Export Along Paths documentation page: https://documentation.samson-connect.net/tutorials/export-along-path/export-atoms-trajectories-along-paths/.
SAMSON and all SAMSON Extensions are free for non-commercial use. To get SAMSON, visit https://www.samson-connect.net.





