Creating analog libraries is a common part of early-stage molecular design or virtual screening workflows. But what happens once you have a list of 2D chemical analogs—how do you go from a SMILES representation to three-dimensional molecular models ready for interaction analysis or docking?
In this post, we’ll look at a lesser-known but powerful feature of the SMILES Manager in SAMSON: the ability to instantly convert your list of 2D analogs to ready-to-analyze 3D molecular structures. This saves time and reduces errors compared to exporting SMILES, using external software to generate 3D geometries, then importing them back into your environment.
Why this matters
As molecular designers, we often need to generate and test structural analogs to assess their potential activity or properties. While it’s easy enough to manipulate SMILES codes, many downstream tasks—such as docking or visual inspection—demand 3D conformations.
If you’re switching between tools to move from 2D to 3D, you’re introducing friction and potential misalignment. The SMILES Manager in SAMSON solves this by keeping everything inside a single interactive modeling platform.
Context: Working with analogs
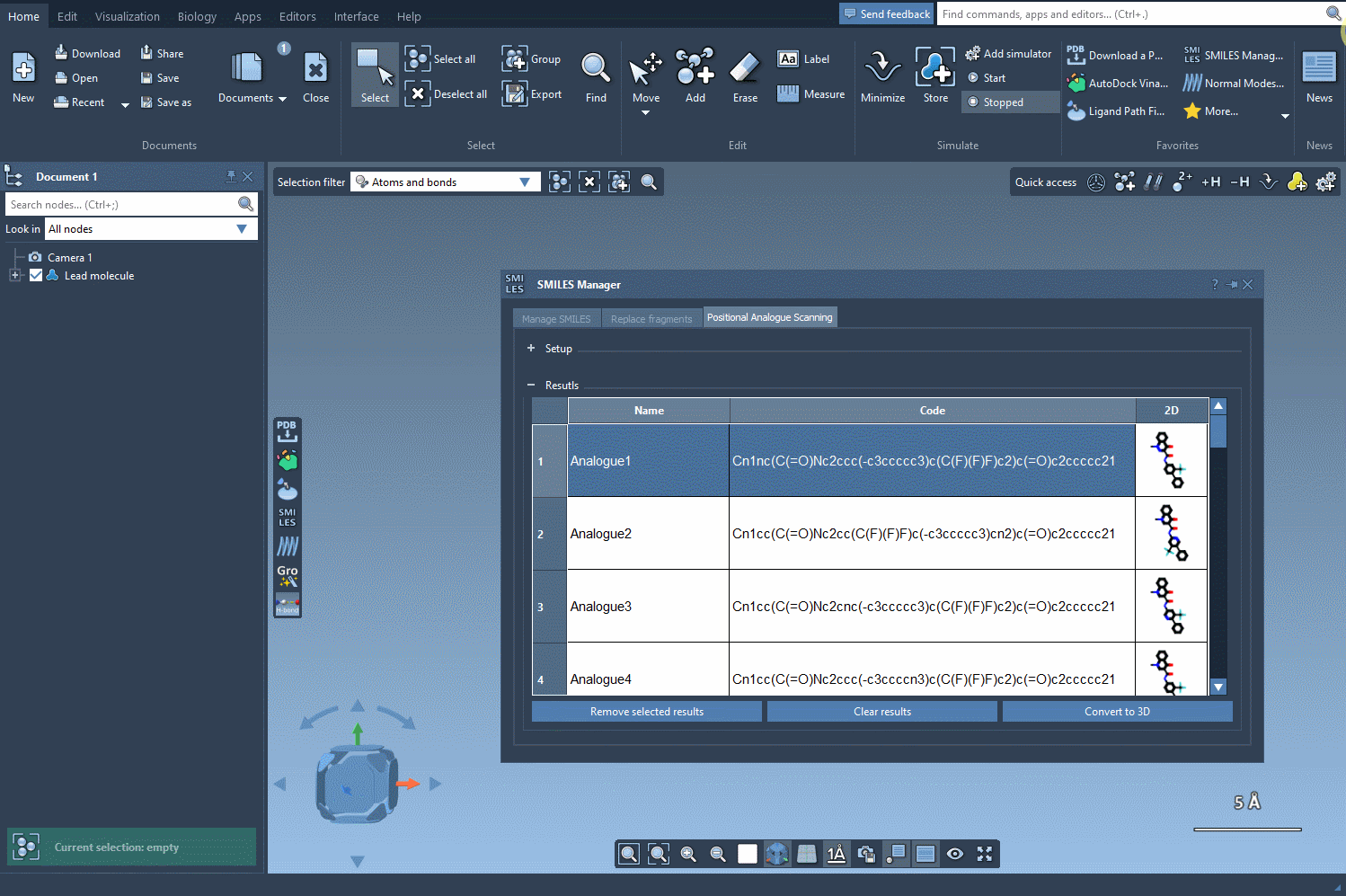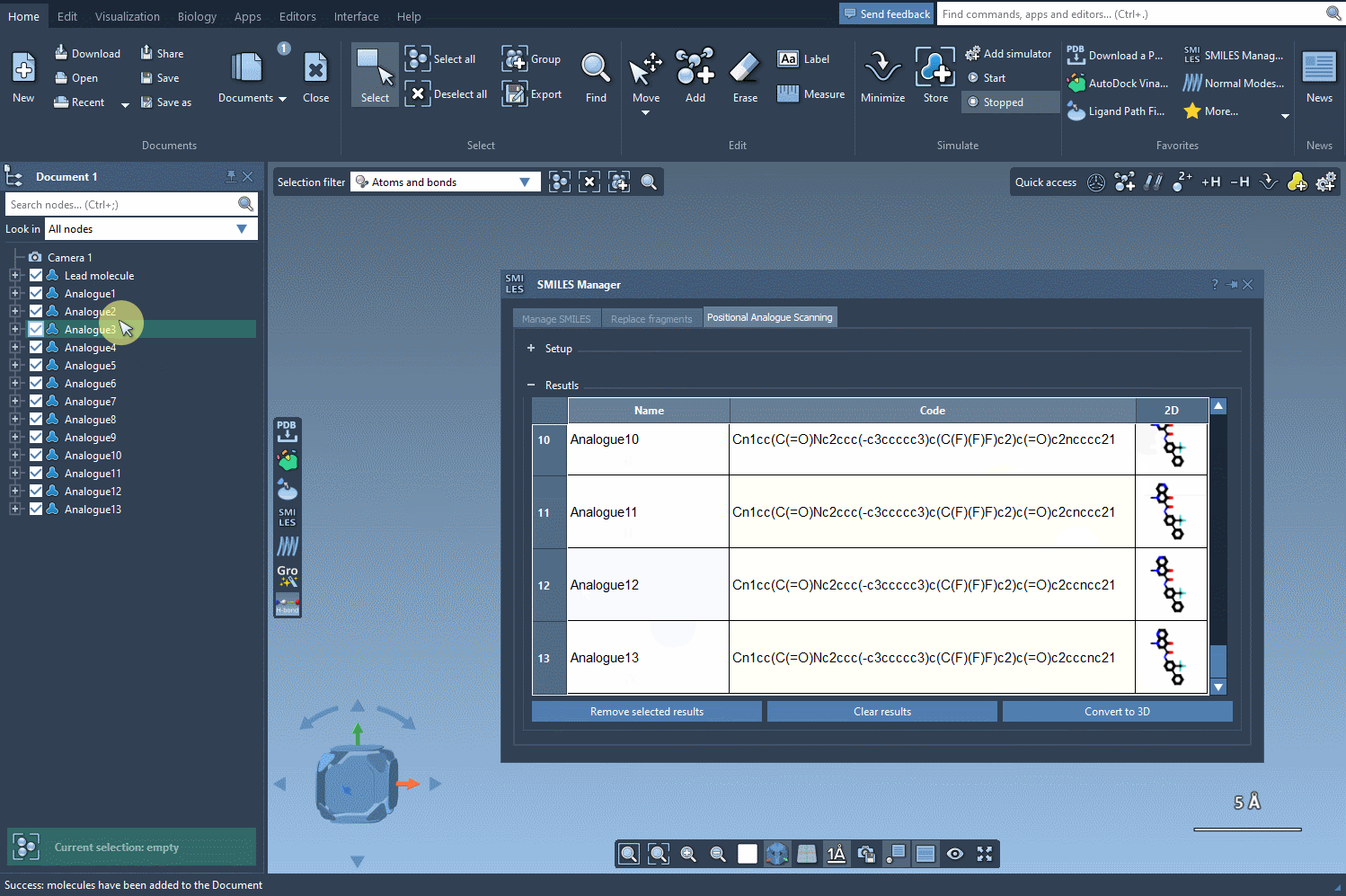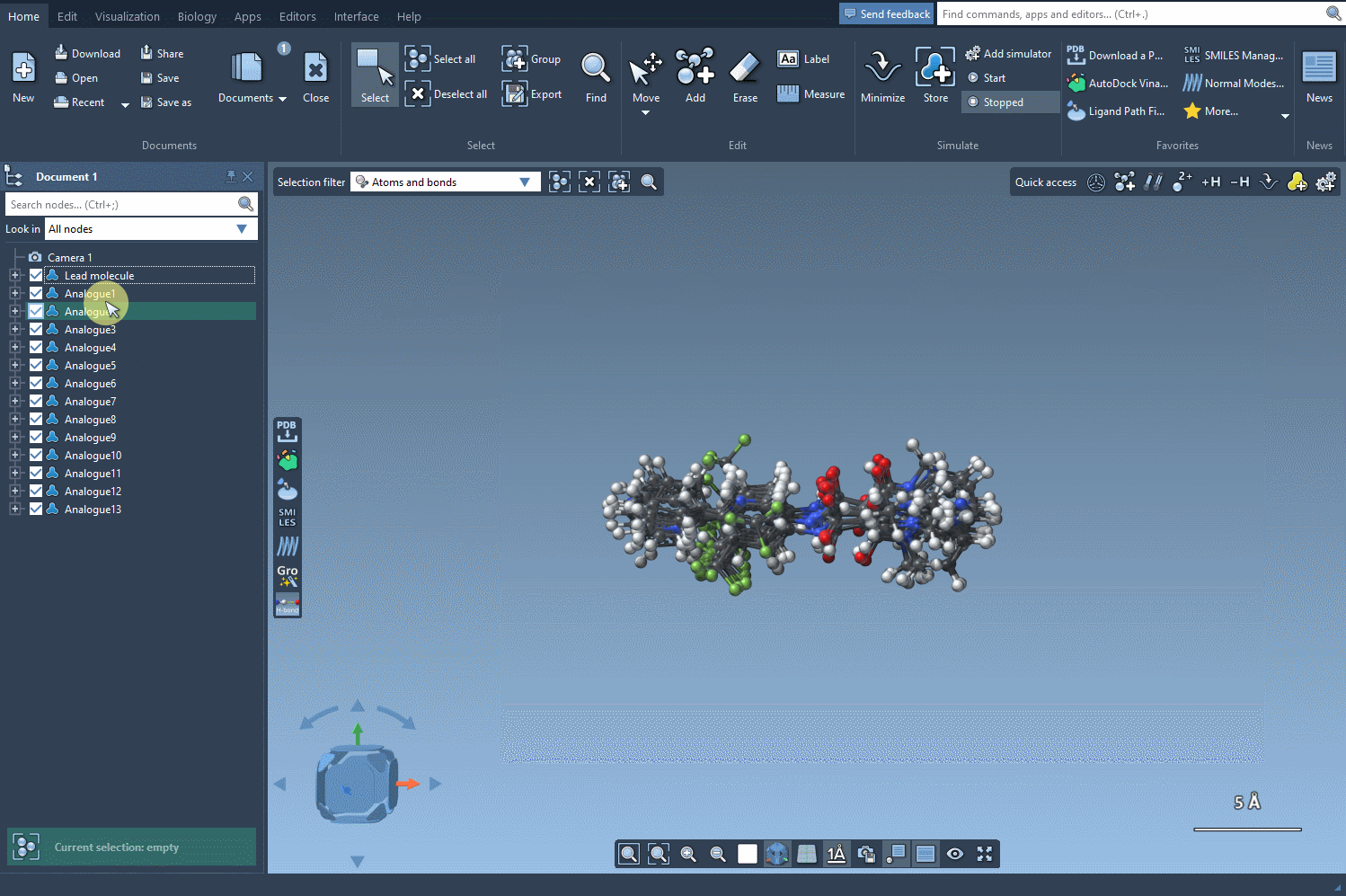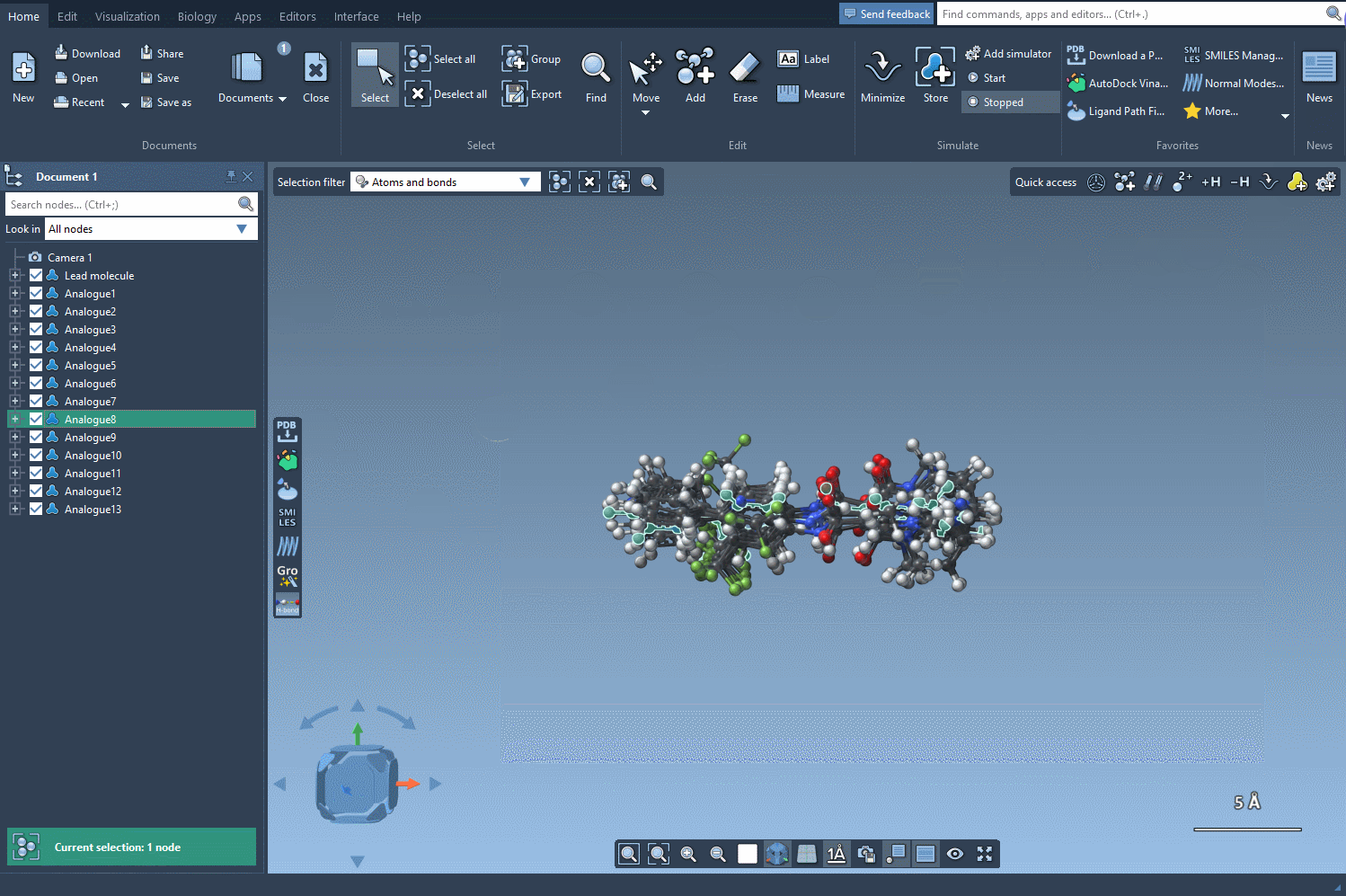
Let’s assume you’ve generated a focused set of analogs using positional analogue scanning—a technique that allows the substitution or attachment of various atoms at well-defined positions in your molecule.
After defining a SMARTS pattern and generating variations, your analogs are displayed in a table with their SMILES codes and 2D depictions.
Converting to 3D
This is where it gets useful: By clicking on the Convert to 3D button found right in the SMILES Manager interface, the selected analogs are automatically translated into 3D molecular structures. The chemical perception and conformation generation are handled behind the scenes.
What you can do next
Once the 3D models are generated, you can:
- Run a molecular docking study using extensions like Autodock Vina Extended.
- Measure distances and angles, or compute interaction energies.
- Visualize how small structural changes affect molecular shape or binding orientations.
A streamlined workflow
This eliminates the need to juggle multiple software tools just to get from a SMILES string to a 3D representation. You stay inside SAMSON, where you can immediately apply additional analyses such as docking, quantum calculations, or visual exploration of the binding site.
In short, this feature helps you quickly go from ideas to models—with fewer clicks and fewer manual steps.
Learn more about this functionality and the full Positional Analogue Scanning workflow in the full documentation: https://documentation.samson-connect.net/tutorials/smiles-manager/perform-positional-analogue-scanning-using-the-smiles-manager-element/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





