When exploring new molecules in drug discovery or materials science, seeing the molecule in 3D isn’t a luxury—it’s a necessity. However, for many molecular modelers, converting a generated 2D molecular structure (e.g., from a SMILES code) into its 3D counterpart can be tedious and time-consuming. What if you could do this in just a few clicks, without jumping between tools?
With the SMILES Manager in SAMSON, transforming 2D analogs into 3D structures becomes a part of your workflow—no need for external file conversions or tool switching. This is particularly helpful when performing positional analogue scanning, a technique commonly used to explore the impact of small changes in a molecular scaffold. After generating analogs through atom replacement or attachment, visualizing their 3D conformations helps researchers assess spatial fit, build intuition about interactions, or prepare for docking simulations.
Why Convert to 3D?
Seeing analogs in 2D gives you a rough idea of their structure, but it leaves out key spatial details like bond angles, steric factors, and the overall three-dimensional molecular shape—elements essential for understanding biological activity or materials behavior. Working in 3D can help:
- Predict binding modes with target proteins
- Evaluate steric clashes or favorable interactions
- Generate more accurate input for docking or simulation tools
How to Generate 3D Structures with SMILES Manager
Once you’ve used the SMILES Manager to generate a set of analogs (e.g., by replacing a hydrogen with a nitrogen atom or attaching a methyl group), generating their 3D structures is a one-step process. Simply click the Convert to 3D button in the interface.
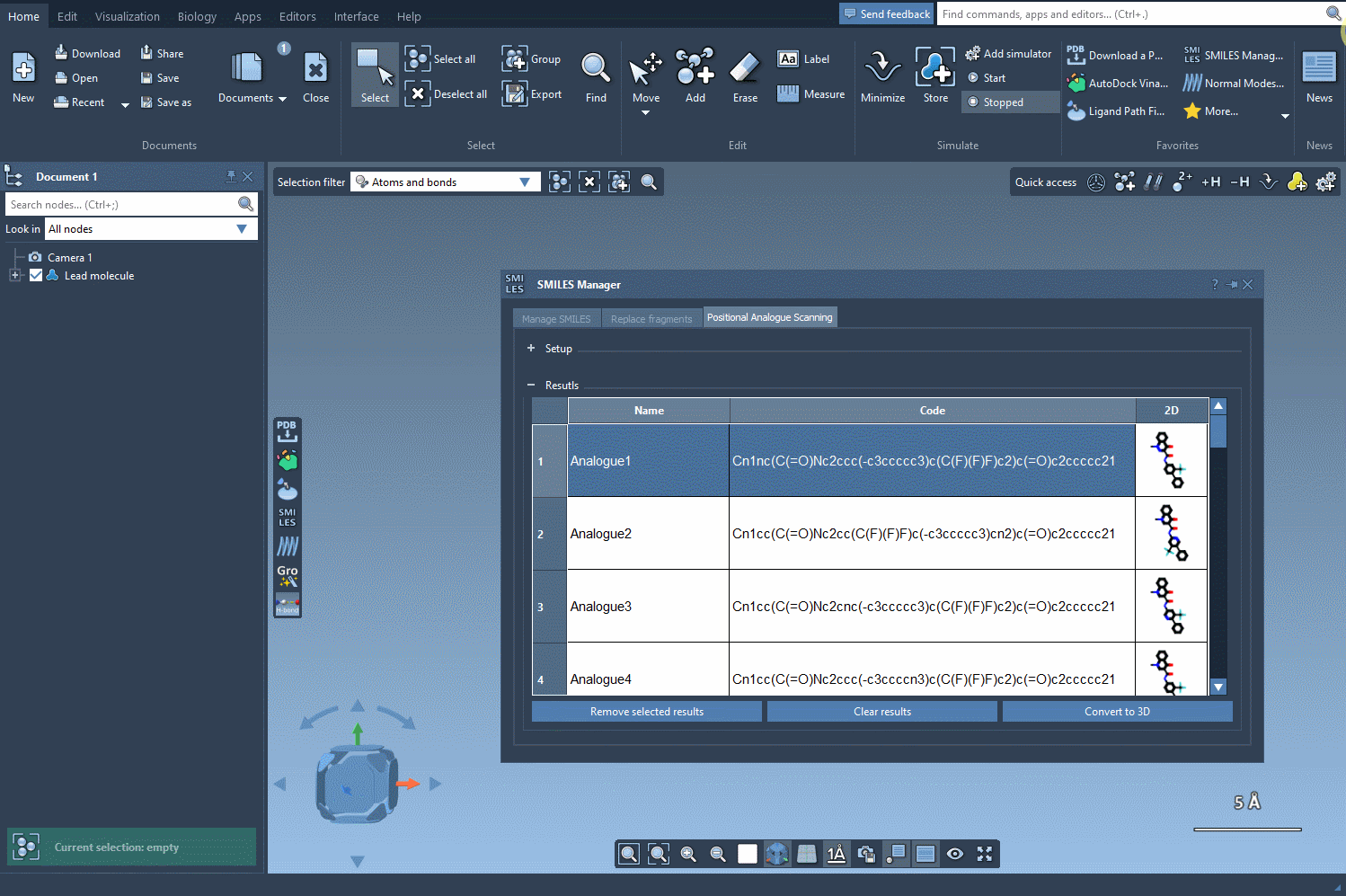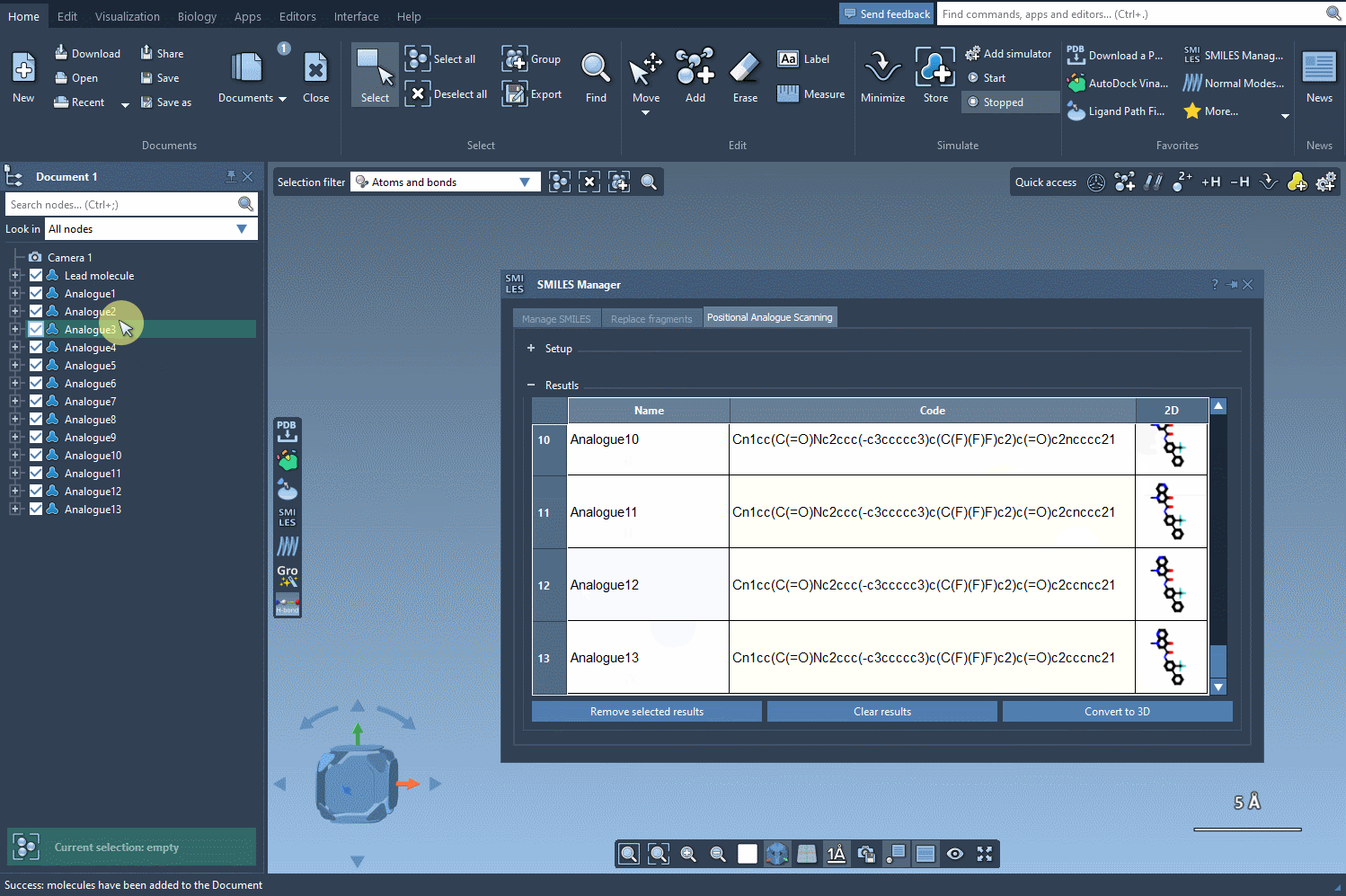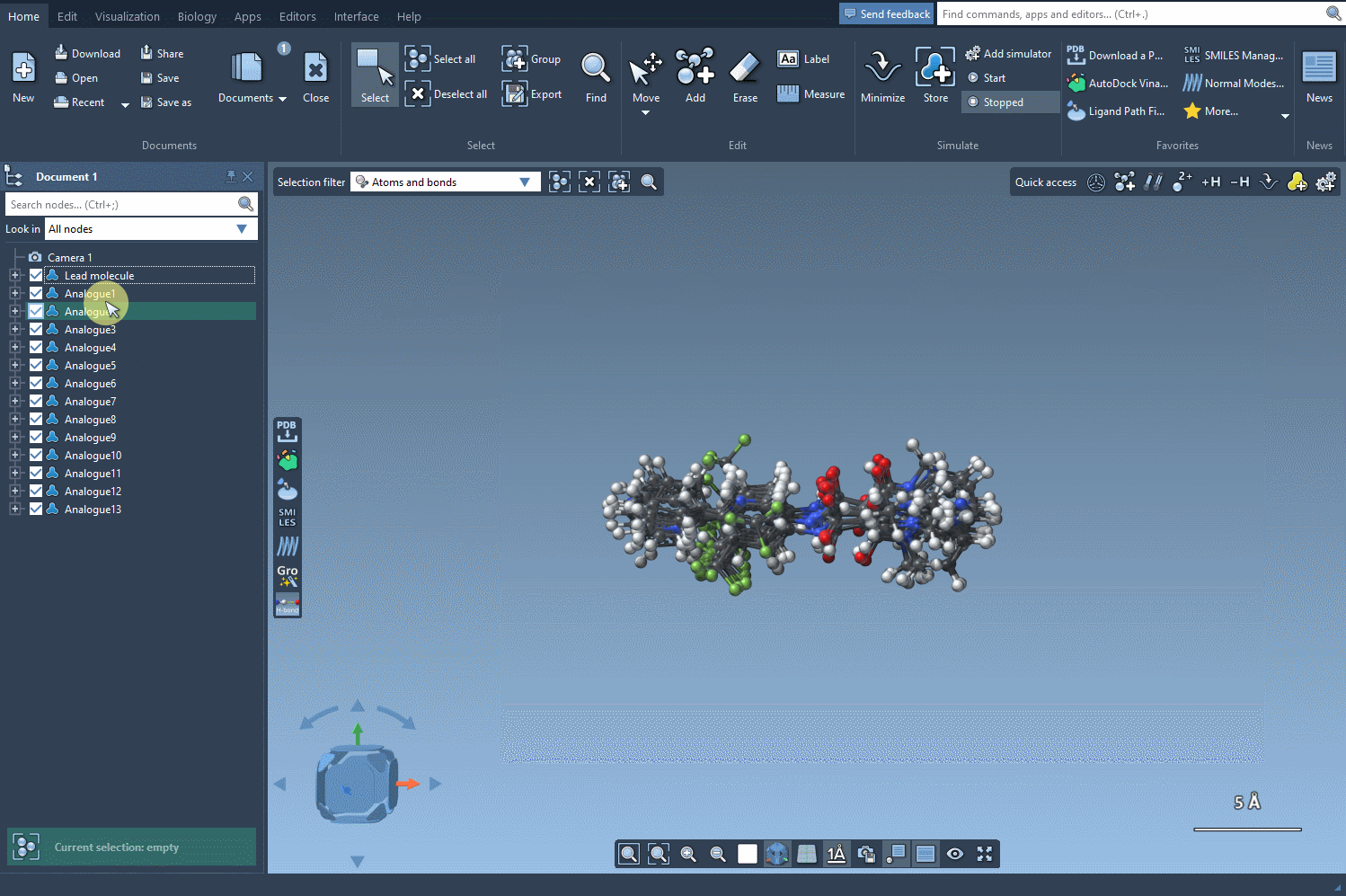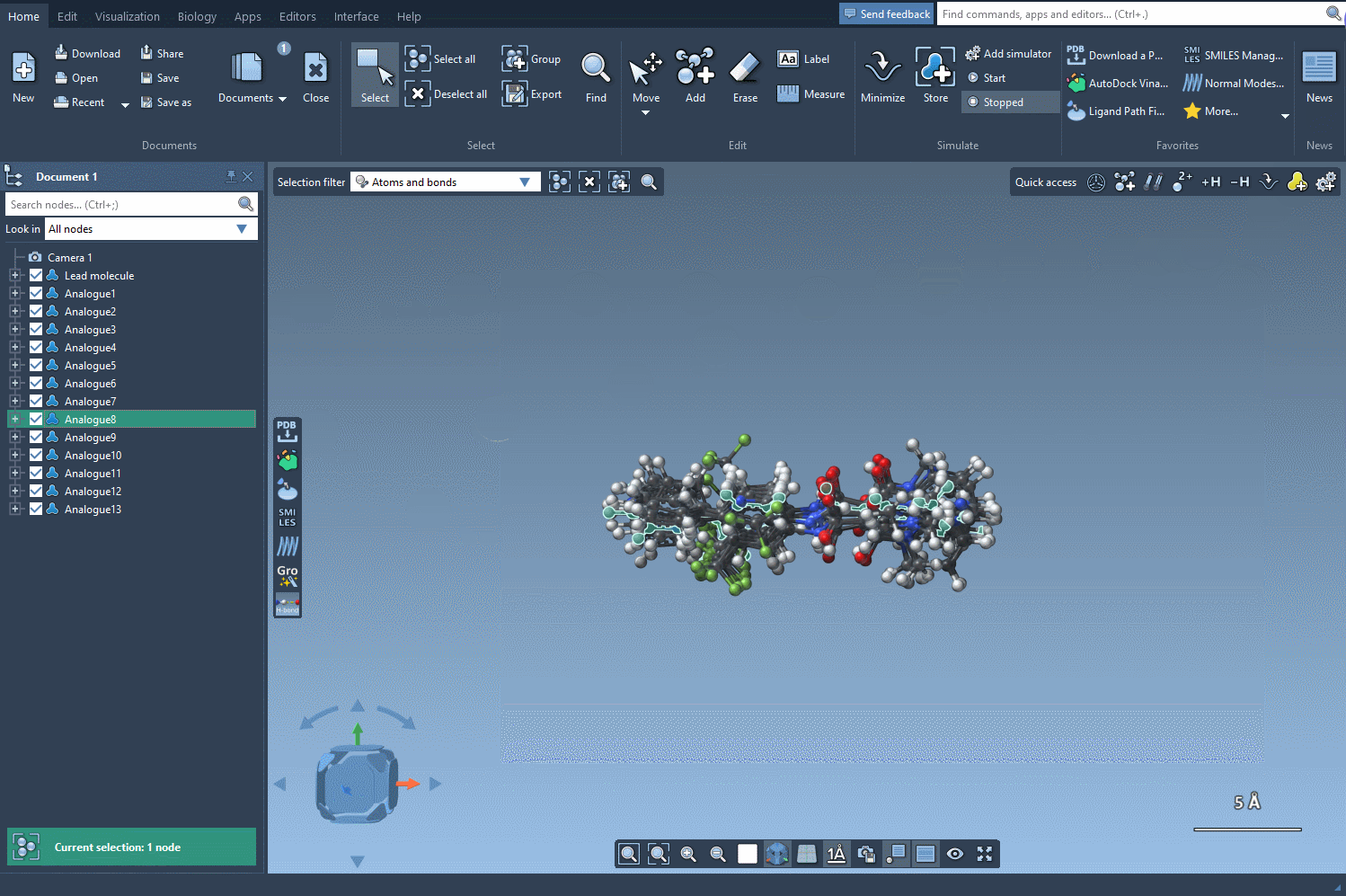
The SMILES Manager will build 3D conformations for each analog and insert them directly into your SAMSON document. Now you’re ready to visually inspect or run further calculations like docking with Autodock Vina Extended.
Tips for Efficient 3D Analysis
- Use the 3D view in SAMSON to compare analogs side-by-side and infer which modifications impact molecular flexibility or spatial arrangement.
- Try coloring atoms or using label annotations to keep track of key changes across analogs.
- Use SAMSON’s selection tools to focus on specific regions of an analog for closer inspection or simulations.
In one interface, the SMILES Manager lets you go from idea to 3D structure in minutes—an integrated path from molecular intuition to visualization.
Want to learn the full process from starting molecule to 3D analysis with positional analogue scanning? Explore the full tutorial at this link.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





