When designing analogs of a molecule in a hit optimization campaign, it’s common to work primarily with 2D molecular structures. But eventually, structural biologists and molecular modelers face an essential question: how do these analogs behave in 3D? Understanding spatial arrangement is critical, especially when evaluating interactions with a target protein or performing docking studies later on.
Fortunately, the SMILES Manager extension in SAMSON offers a way to quickly convert your generated 2D analogs into 3D structures and analyze them in your modeling workflows.
Why 3D Structures Matter
2D analog designs are often the starting point of brainstorming molecule variants. But conformation changes, steric clashes, or spatial rearrangements can determine whether a molecule binds tightly to its target—something 2D structures simply can’t reveal.
That’s what makes the Convert to 3D feature of the SMILES Manager extension so useful: it lets you examine the resulting conformations right after analogue generation.
How Does It Work?
Once you’ve generated a set of analogs through positional analogue scanning, converting them to 3D in SAMSON just takes a click. Click on the Convert to 3D button inside the SMILES Manager interface, and all analogs are processed to output 3D molecular geometries that you can directly visualize and manipulate in the SAMSON workspace.
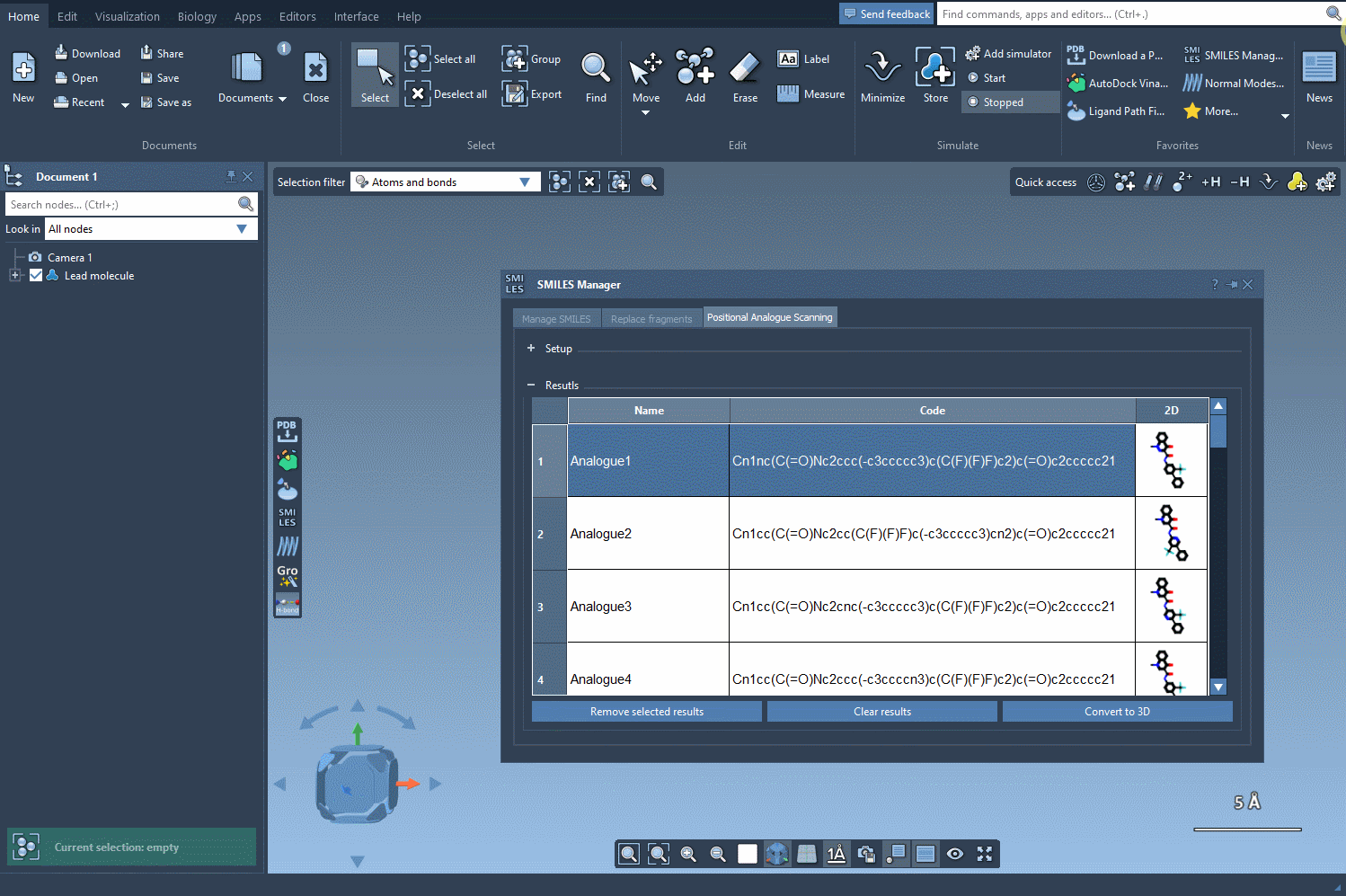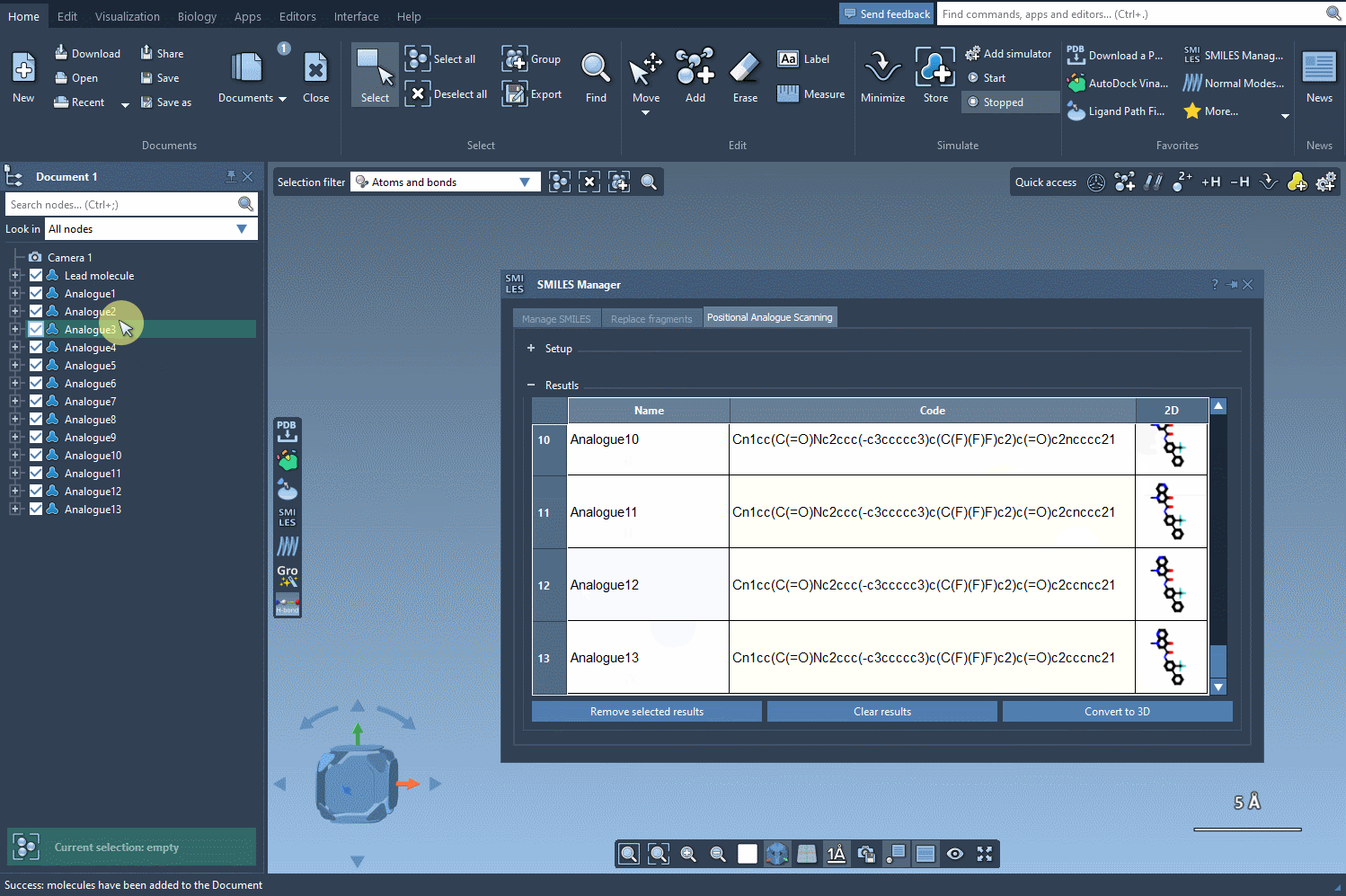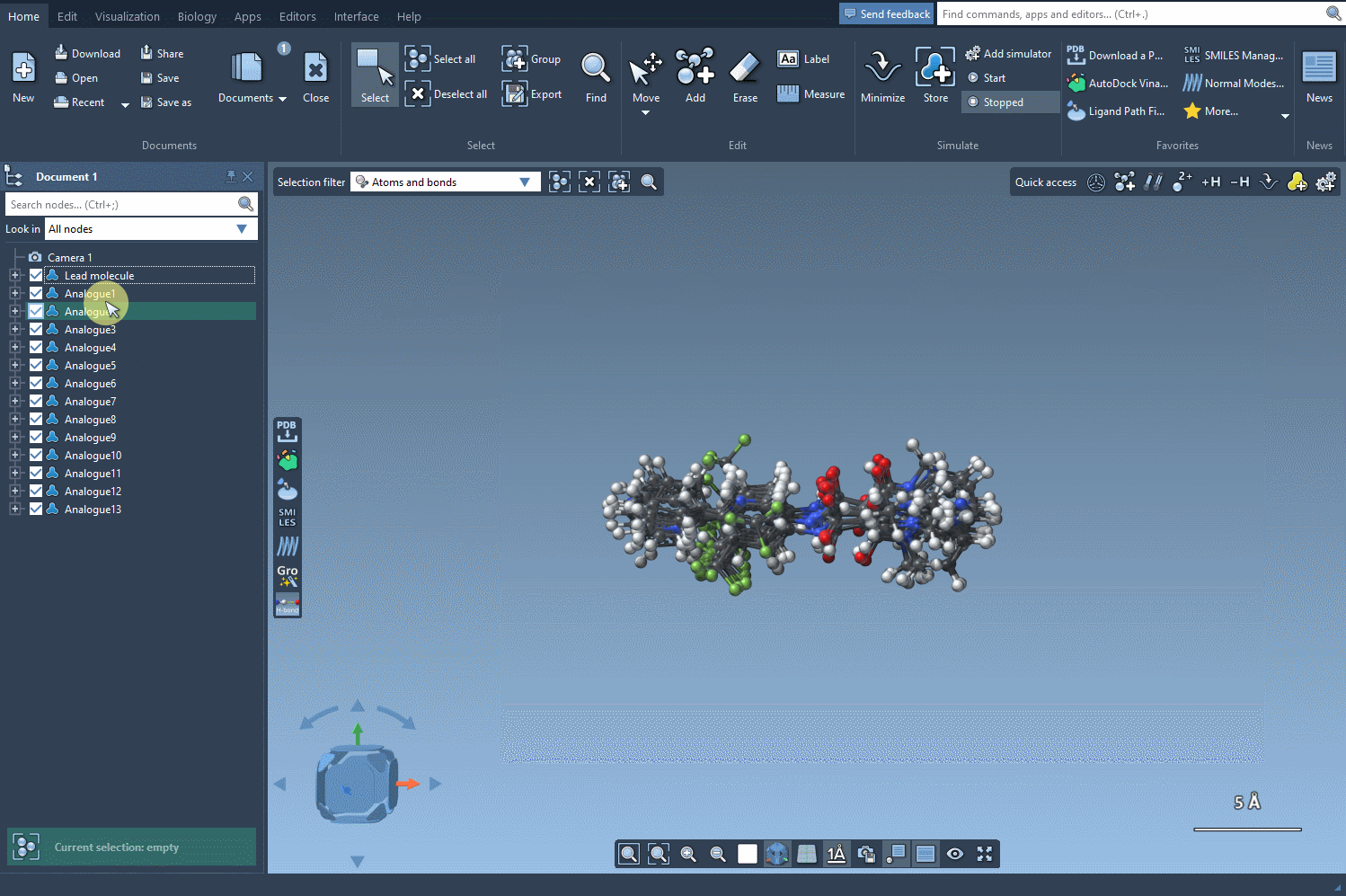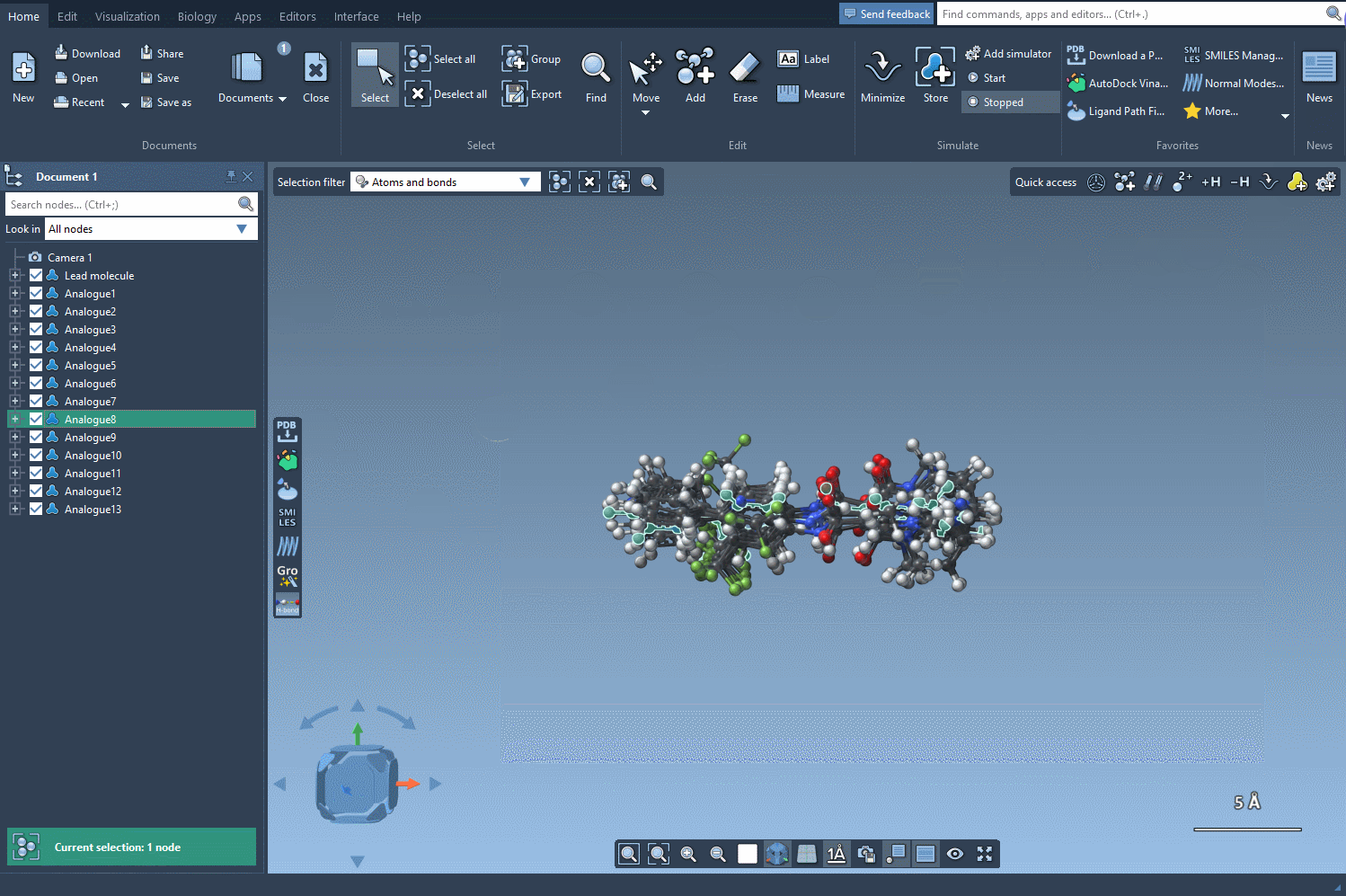
What Can You Do With The 3D Conformations?
- Perform Docking: Analyze how variations impact protein-ligand interactions using the Autodock Vina Extended extension.
- Visual Inspection: Check whether added groups introduce strain or if new hydrogen bonds or hydrophobic contacts are created.
- Compare Analogs: With all variants loaded in 3D, you can quickly spot which modifications lead to favorable conformations and which may be problematic.
This step essentially bridges the gap between fast 2D analogue generation and advanced computer-aided drug design decisions.
Streamline Your Pipeline
Many researchers use external tools to simulate or generate 3D conformations from SMILES strings. This can involve converting files, importing into other programs, and sometimes losing annotations in the process. By contrast, with SAMSON, all analogs generated from a positional scan stay within the same project and environment—and can be immediately studied in their 3D form.
Conclusion
Whether you’re quickly exploring SAR ideas or preparing inputs for docking, being able to generate and analyze 3D analog structures directly in SAMSON is a time-saving and practical feature. Each button click brings you closer to chemically meaningful insights about your molecule set.
To learn more about the full analogue generation process using the SMILES Manager, check out the complete tutorial here: https://documentation.samson-connect.net/tutorials/smiles-manager/perform-positional-analogue-scanning-using-the-smiles-manager-element/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





