Finding reliable transition pathways between molecular conformations is a central task in computational chemistry and molecular modeling—whether for studying protein-ligand binding, enzymatic mechanisms, or conformational changes. The Nudged Elastic Band (NEB) method helps compute minimum energy paths (MEPs) between known states of a system, and the Parallel Nudged Elastic Band (P-NEB) implementation in SAMSON makes this easier and faster. However, many users wonder: Should I run P-NEB on my set of conformations or convert them into a path first?
This post focuses specifically on that choice, aiming to save you both time and computational resources while simplifying your workflow. If you’ve collected or generated conformations representing an unbinding or reaction path, read on.
Paths vs. Conformations: What’s the Difference?
In SAMSON, conformations are nodes that store atom positions from a particular state. These can be saved snapshots from an MD trajectory, user-generated states, or interpolated structures.
On the other hand, a path is a container for a trajectory—essentially a connected series of conformations with a notion of directionality. It can be animated and interacted with. Importantly, the P-NEB app can optimize both—but not equally efficiently.
Performance Matters: Prefer Paths
If you’re deciding whether to run P-NEB on conformations or convert them into a path first, the recommendation is clear: prefer using a path.
Note
For the same system and number of intermediate states, applying P-NEB to a path will be faster and more memory-efficient than applying it directly to a set of conformations.
To convert conformations into a path, select them in the Document view, right-click, and choose Conformation > Create path from conformations.
Why It Works Better
When using a path input, the NEB app can streamline its access to data, better handle interpolation between intermediate states, and benefit from animation and visualization tools in SAMSON. This facilitates both setup and interpretation. In contrast, raw conformations require additional overhead for indexing, ordering, and tracking during optimization.
Visual Workflow Example
For instance, if you have a ligand unbinding trajectory saved as a sequence of conformations, you can combine them and apply P-NEB to the resulting path:
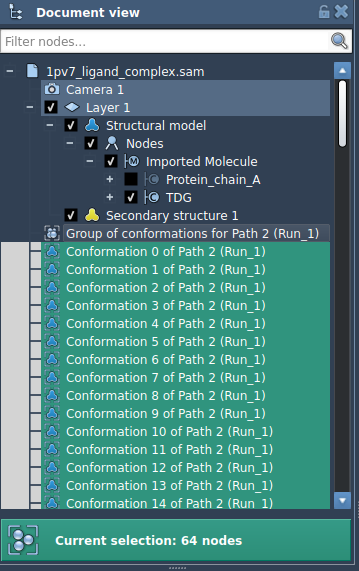
But after converting them into a path, the optimization is smoother and faster:
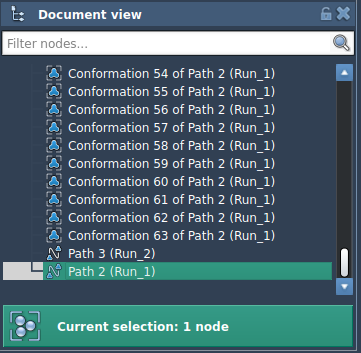
The output is also clearly organized in the Document view as a new optimized path:
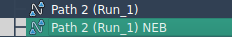
Conclusion
For those frequently working with multiple conformations in transition studies, it’s worth building the habit of grouping them into a path before running P-NEB. It saves time and often improves results. To learn more and explore the full tutorial, including interactive settings and sample projects, visit the official documentation:
documentation.samson-connect.net/tutorials/pneb/…
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at www.samson-connect.net.





