Designing specific polymer architectures often starts with one fundamental step: defining your own monomers. For molecular modelers working on custom polymers, functionalized scaffolds, or bioconjugates, creating a library of reusable, well-defined repeat units can streamline workflows and minimize errors later in the chain.
If you’re using SAMSON, the integrative molecular design platform, the Polymer Builder extension includes an intuitive monomer registration step. This post walks you through how to register, edit, and organize custom monomers, and why this step is worth getting right from the start.
Why Register Monomers?
Whether you’re designing synthetic polymers or biopolymers, having named, clearly defined building blocks helps you:
- Re-use fragments consistently across different projects
- Build sequences with controllable connectivity and bond types
- Predict structural metrics like molecular weight and chain length
Step-by-Step: Registering a Monomer
Here’s how to define a custom monomer starting from a selected molecule in SAMSON:
- Select a molecule (a monomer unit) in the Document view or the Viewport.
- Click Register monomer from selection.
The Polymer Builder will automatically determine the start and end atoms—usually at opposite ends of the molecule—so the monomer can connect cleanly to others. If the automatic choice doesn’t suit your purpose, you can override it:
- Use
Sto select the atom manually based on your current selection. - Use
Pto pick from a list of atoms. - Use
Vto visually highlight atoms or structures.
Each monomer gets a unique letter (e.g., A, B, C…), which becomes its identifier in later sequences.
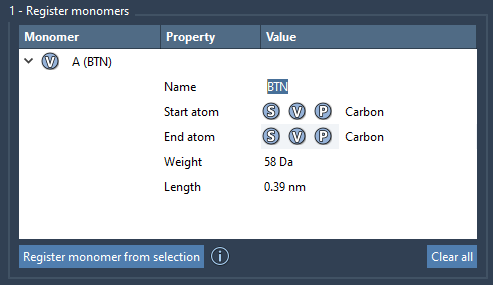
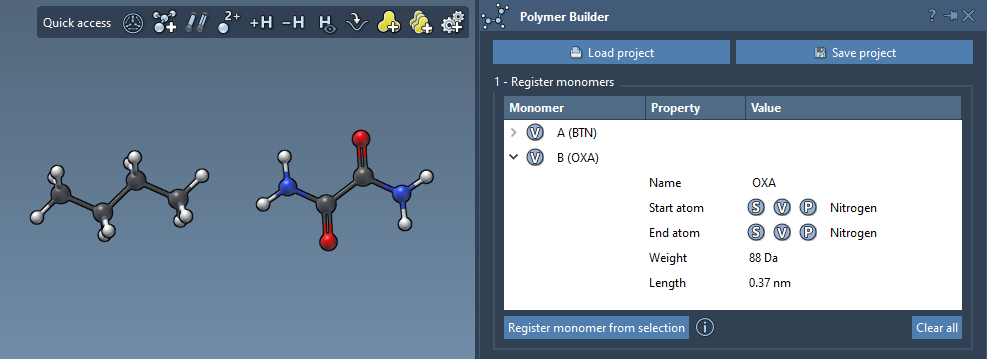
Behind the Table
Once registered, monomers appear in a table that shows:
- Molecular weight (in Daltons)
- Distance between start and end atoms
- Monomer name (automatically retrieved or user-defined)
You can rename or edit monomers directly in the table. For example, you might want to organize monomers into structural groups, useful when rendering or analyzing assembled polymers.
You’ll see automatic feedback if parts of your monomer have been removed or altered—when a registered molecule is modified, it will automatically be deregistered to avoid inconsistencies.
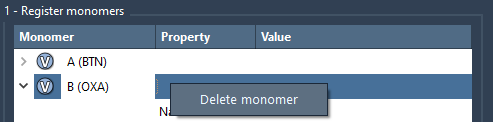
Cleaning Up
If you no longer need a monomer, just right-click on it in the table and select Delete monomer. You can also clear your full list of registered monomers with Clear all.
Tips for Better Results
- If your molecule consists of a single residue or group, its name will be auto-filled when registering. Otherwise, assign a name manually to help differentiate it later.
- To support branching or uncommon connections, verify start and end atoms precisely.
- You can use structures generated from Polymer Builder as new monomers to build block copolymers.
With your monomers registered, you can move on to building sequences or directly generating complex polymers. Starting with a clean, curated monomer panel sets the stage for smoother modeling downstream.
To learn more about Polymer Builder and related features, visit the official documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





