One common challenge molecular modelers face when building polymers is controlling the exact sequence of monomers. Whether you’re designing a repeating synthetic chain or trying to mimic the behavior of a biopolymer, sequence matters. And yet, defining and managing specific patterns of monomers in molecular modeling tools can often feel more cumbersome than creative.
The Polymer Builder extension for SAMSON addresses this problem with sequence registration and editing tools made for modelers who need precision without extra friction. If you’ve already registered your custom monomers, this functionality allows you to bundle those units into structured sequences that define exactly how your polymer chain will form — including the type of chemical bonds joining each unit.
Define Sequences with Clarity
Once you’ve registered individual monomers, the next step is to define the logic: how monomers will be repeated and ordered. The “Register Monomer Sequences” function in Polymer Builder simplifies this into a few intuitive steps:
- Click Add new sequence.
- Name the sequence and define its structure using monomer IDs (e.g.,
ABBA).
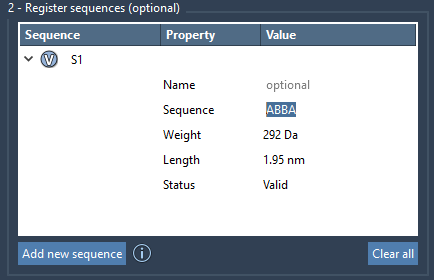
Sequences are labeled automatically (S1, S2, etc.), but you can assign them custom names as needed. Additionally, the tool provides helpful features like monomer highlighting (via the V button next to each sequence), and clearly displays validity status or potential sequence errors.
Control Chemical Bonds
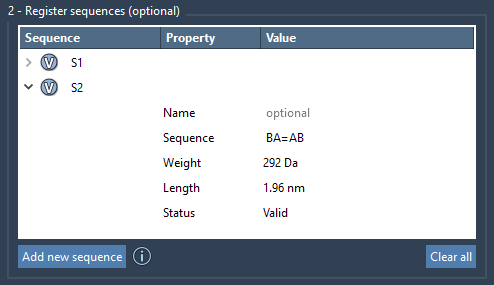
A great feature in this tool is specifying bond types directly in the sequence. By default, monomers connect via single bonds. But if your structure calls for variations, just use:
=for double bonds (e.g.,A=B)#for triple bonds (e.g.,A#B)
This enables fast prototyping of conjugated polymers or polymeric systems where connectivity influences electronic or mechanical outcomes.
Preview, Edit, and Organize
Each registered sequence is listed in a table that shows:
- The sequence name
- The string pattern (e.g.,
BAAB) - An estimate of molecular weight and length
- Its validation state
You can edit sequences on the fly: update their names, adjust the monomer order, or change bonding types. If a sequence proves problematic, right-click it to remove, or use “Clear all” to start fresh.
Why This Matters
For tasks involving structure-property prediction, efficient simulation setup, or materials informatics, controlling monomer sequences isn’t just helpful — it’s essential. Whether you’re modeling drug-polymer conjugates or experimenting with copolymer arrangements, sequence design dramatically shapes your system’s behavior. The Polymer Builder’s sequence registration streamlines these needs with minimal overhead.
To learn how this fits into the full workflow — from designing a monomer to generating a complete polymer — visit the official Polymer Builder documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at www.samson-connect.net.





