Designing new analogues of a lead compound is a daily task for many molecular modelers and medicinal chemists. But one recurring challenge remains: how can we efficiently suggest and visualize meaningful changes to a molecule—and study their effect on properties like binding affinity or key molecular interactions—without relying on a complex pipeline of tools?
For those working with SAMSON, there’s a way to scan through potential chemical modifications rapidly using the SMILES Manager extension. One particularly useful feature is the ability to generate 3D structures of analogues directly from SMILES, helping you go from concept to visual analysis in seconds.
A Common Bottleneck
Let’s say you’ve identified a promising molecule. You want to understand how substituting a specific atom—say replacing an aromatic carbon with a nitrogen—might improve interactions with your protein target. You’ve already created several variations, but building their 3D models to test docking or visualize interactions takes time and effort, especially when dealing with large libraries of analogs.
With SAMSON’s SMILES Manager, this process becomes much more straightforward. Here’s how.
Seamlessly Generate 3D Structures
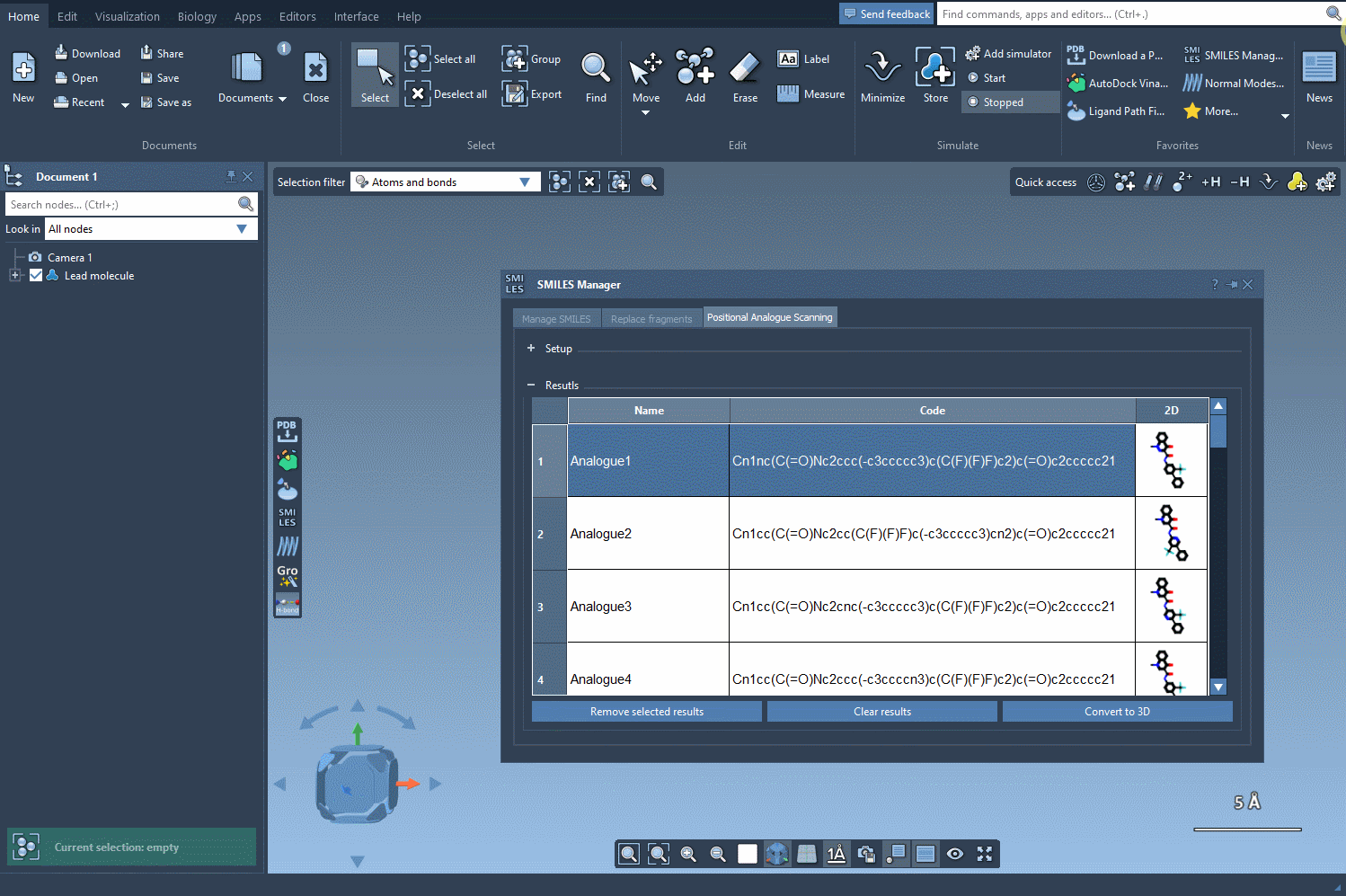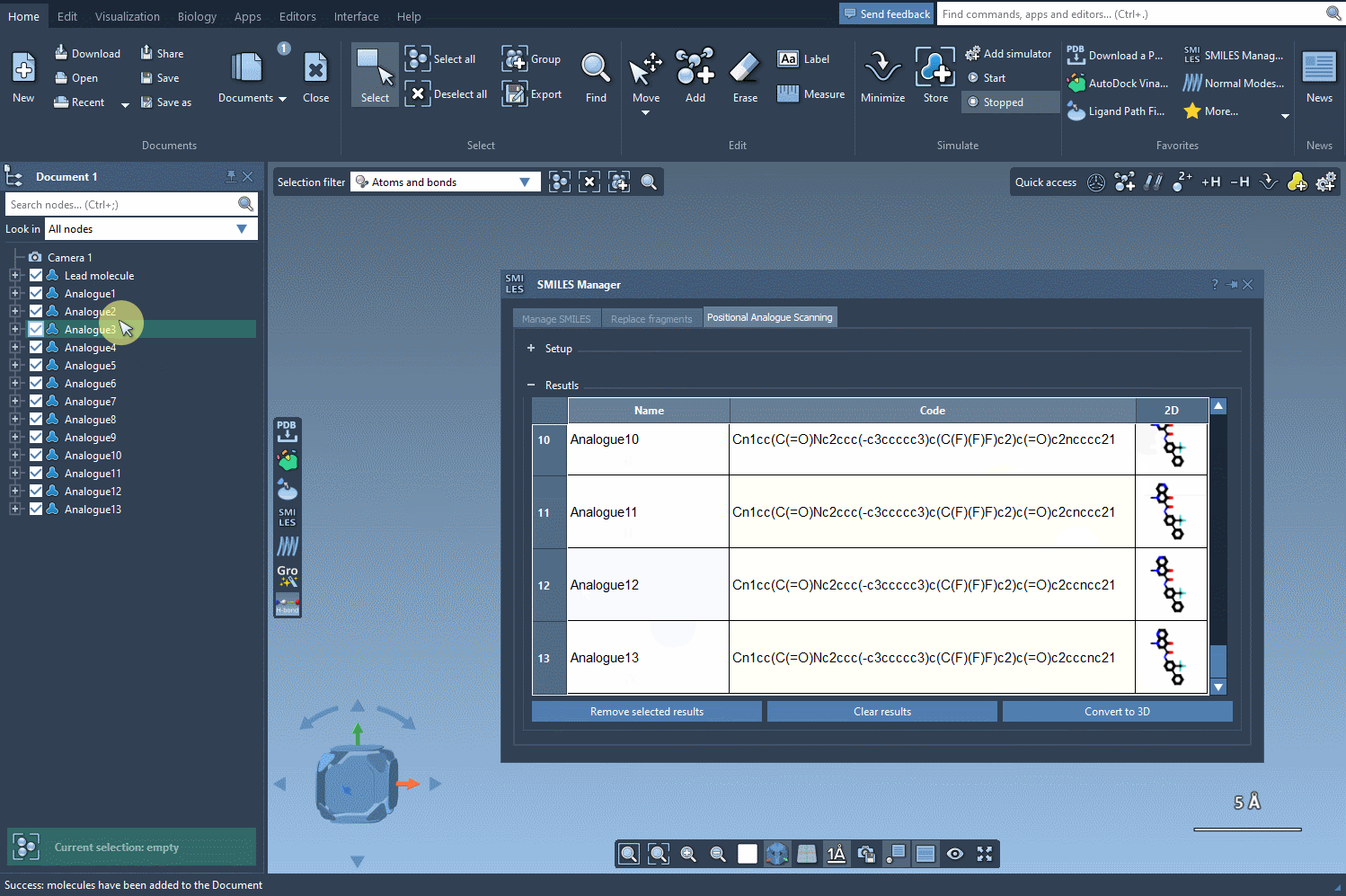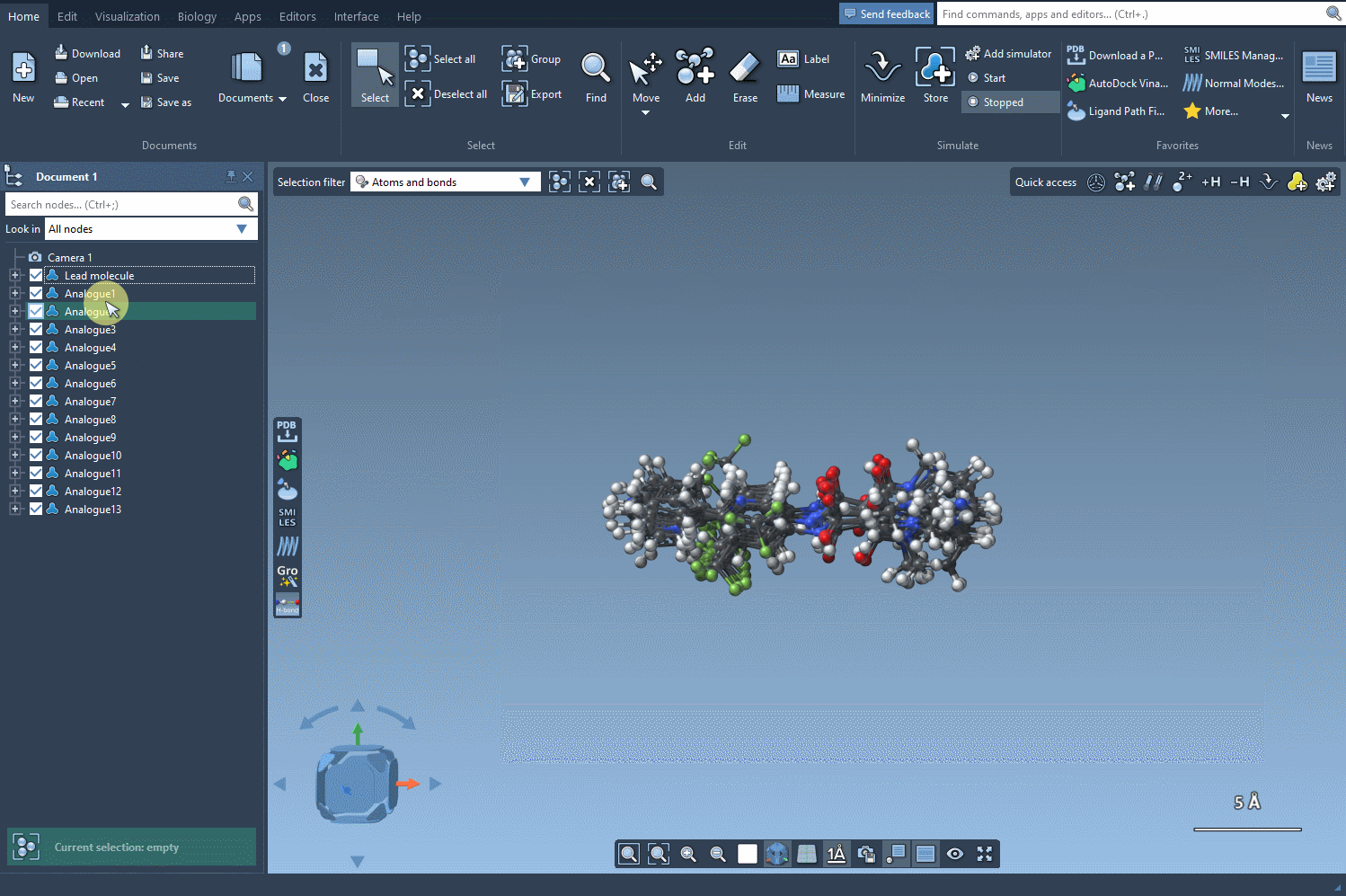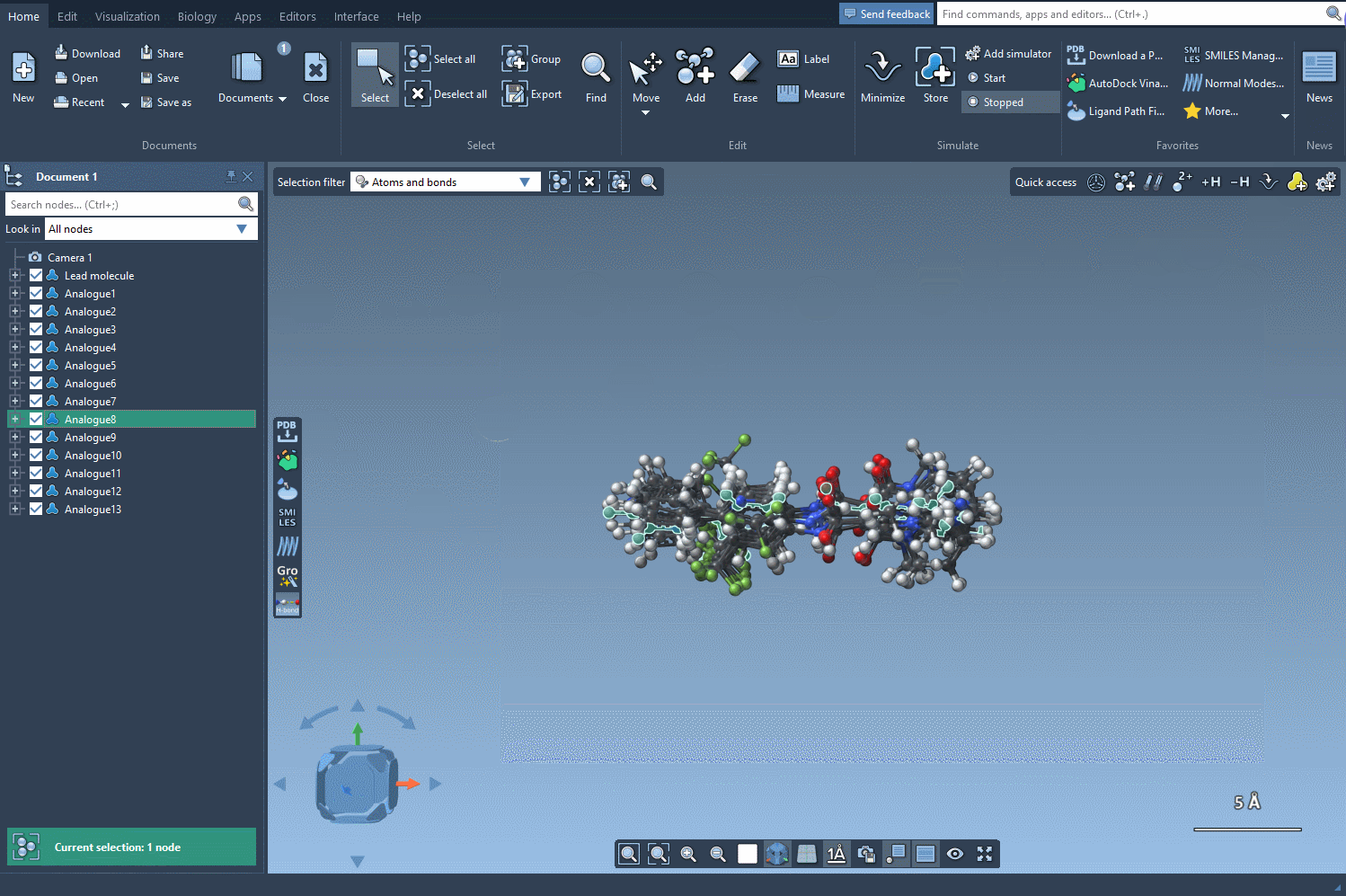
After defining your molecule and identifying the pattern to modify (e.g., using the SMARTS pattern [cH] for an aromatic carbon), you can generate new analogs with modified groups. The analogs are listed in a table, each with a 2D depiction generated automatically.
Now comes the key part: by clicking on Convert to 3D, SAMSON generates and displays 3D structures of the selected analogs. No need to export files or switch between platforms—it all happens within the same environment.
This helps you quickly:
- Visualize how atom substitutions affect molecular geometry
- Prepare the structures for downstream tasks like docking
- Communicate modifications clearly with exportable visuals
What Happens Next?
Once 3D structures are generated, you can dock them using other SAMSON extensions (like Autodock Vina Extended) to evaluate binding affinity. You can also inspect whether new interactions form, using the complete visualization tools integrated into SAMSON’s workspace.
The process is particularly helpful in iterative design workflows where different modifications can be compared quickly to assess which changes are worth pursuing further.
Why It’s Useful
Quickly generating 3D structures removes a common hurdle in molecular modeling workflows. It allows for a more seamless integration between hypothesis (“What if I make this modification?”) and evaluation (“How does it influence binding geometry?”). For chemists and modelers, this means faster iterations and more visual insights into the implications of structural changes.
You can learn more about the entire workflow and other features of the SMILES Manager by visiting the full documentation: https://documentation.samson-connect.net/tutorials/smiles-manager/perform-positional-analogue-scanning-using-the-smiles-manager-element/
SAMSON and all SAMSON Extensions are free for non-commercial use. Get SAMSON at https://www.samson-connect.net.





