One of the recurring challenges in cheminformatics and molecular modeling is converting a simple SMILES string into a realistic 3D molecular structure. For many modelers, this means juggling scripts, toolkits, or web services — often repeating the same task across dozens or even hundreds of molecules.
If you’ve faced this, the SMILES Manager extension in SAMSON offers a fast, integrated solution that simplifies 3D structure generation from SMILES — batch or individually — without writing code.
Why Generate 3D Structures from SMILES?
SMILES (Simplified Molecular Input Line Entry System) strings are compact and human-readable molecular representations, but they lack spatial information. For simulations, docking, or structure-based design, you need accurate 3D geometries. That’s where RDKit-based tools like the SMILES Manager come in.
How It Works in SAMSON
The SMILES Manager in SAMSON offers a clean and interactive interface to manage, view, and convert SMILES strings into 3D structures. Here’s how to use its 3D generation feature:
Step 1: Import SMILES Strings
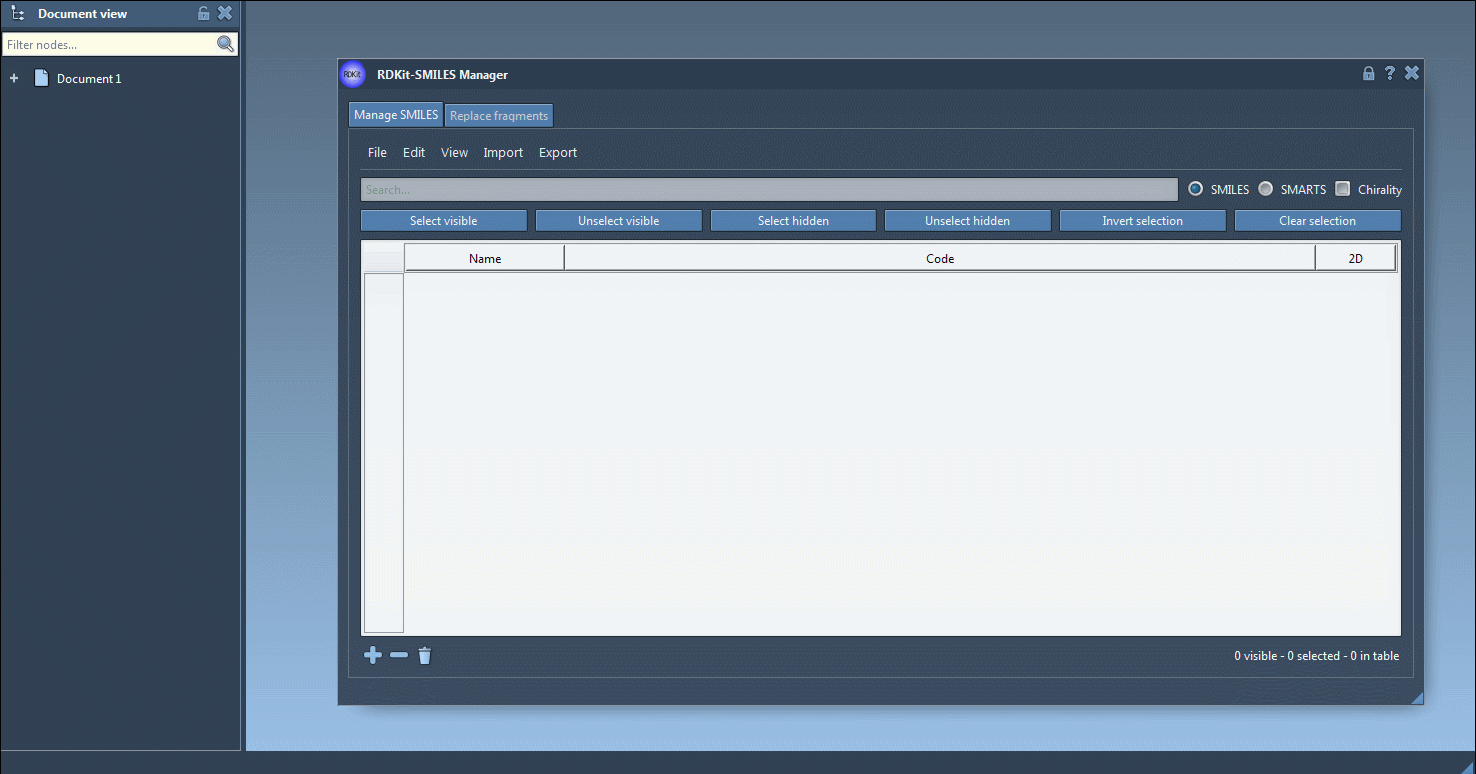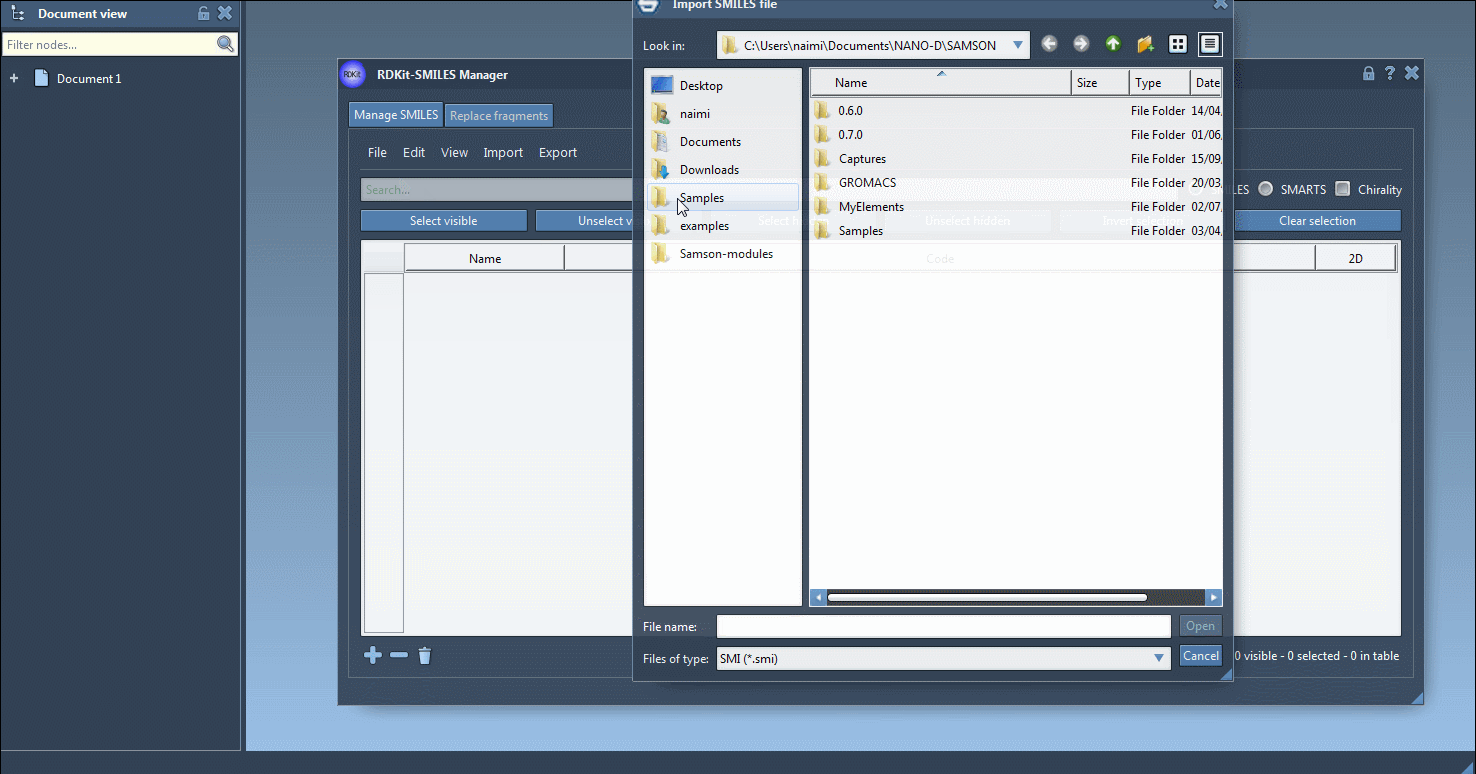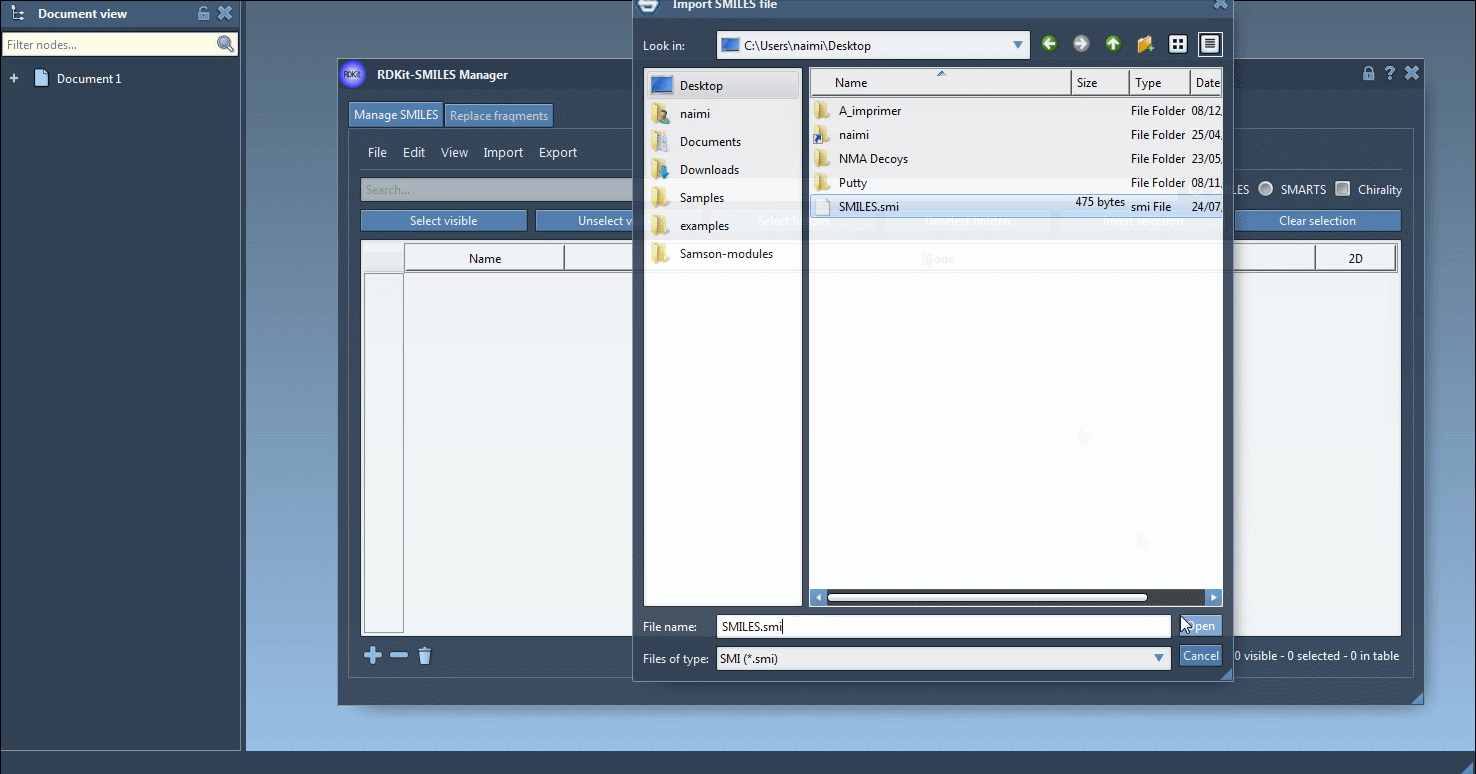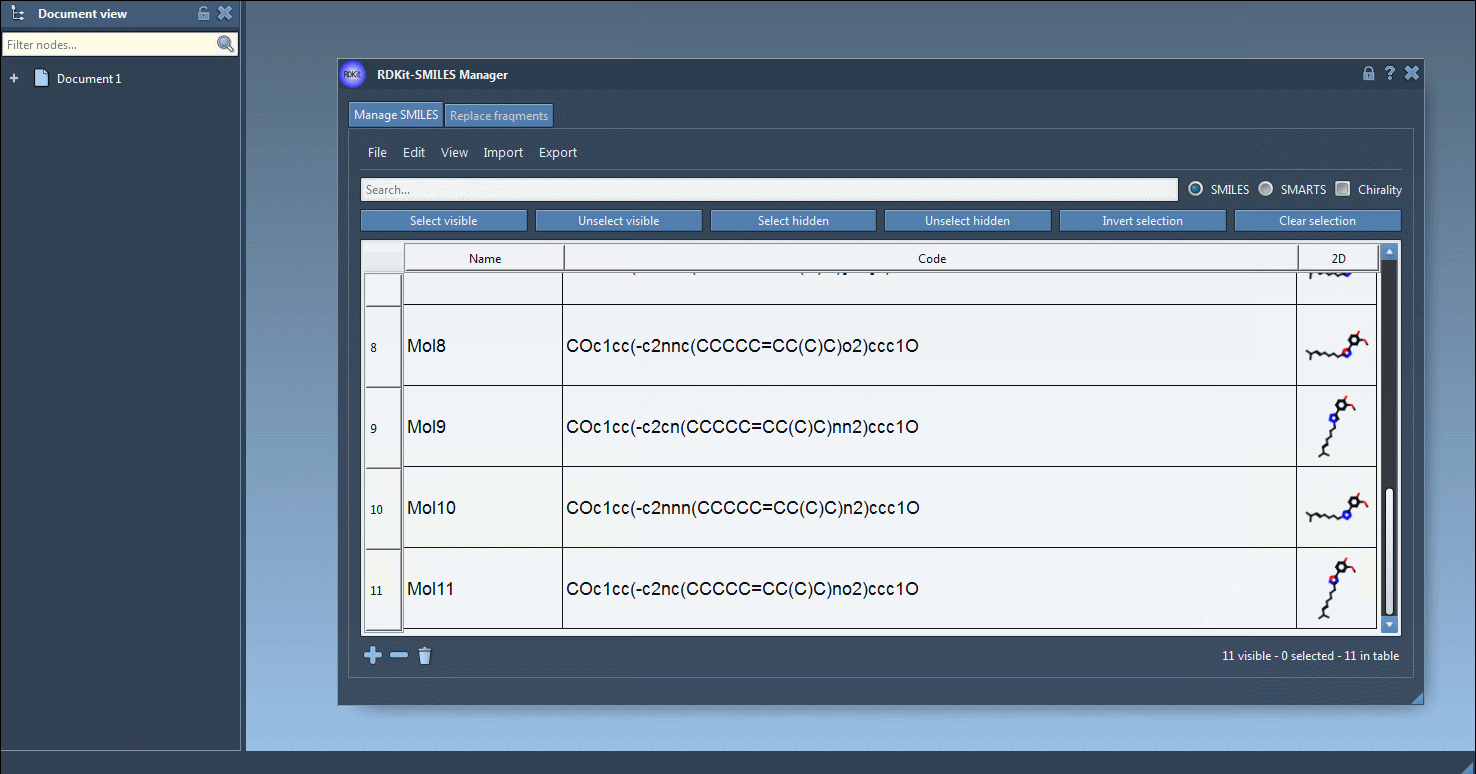
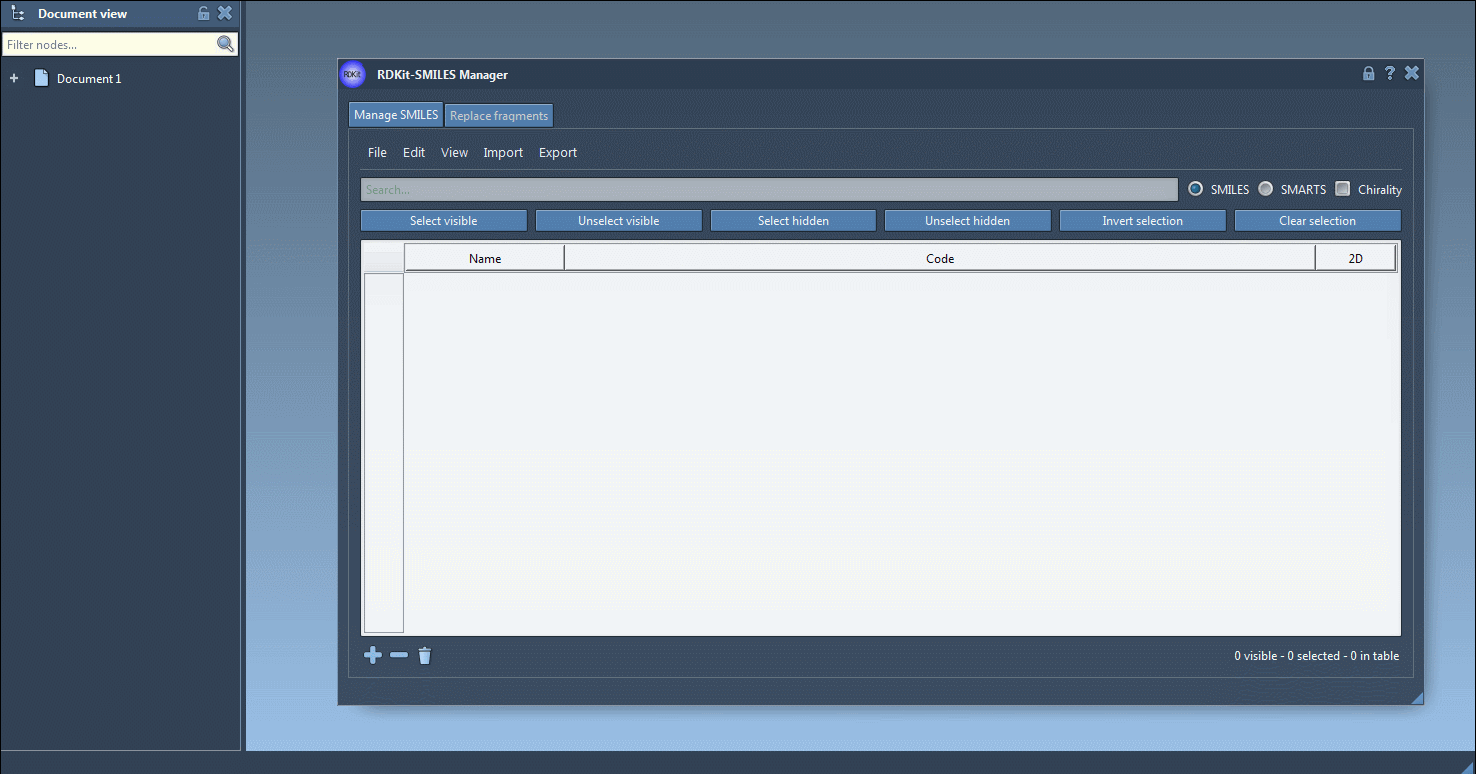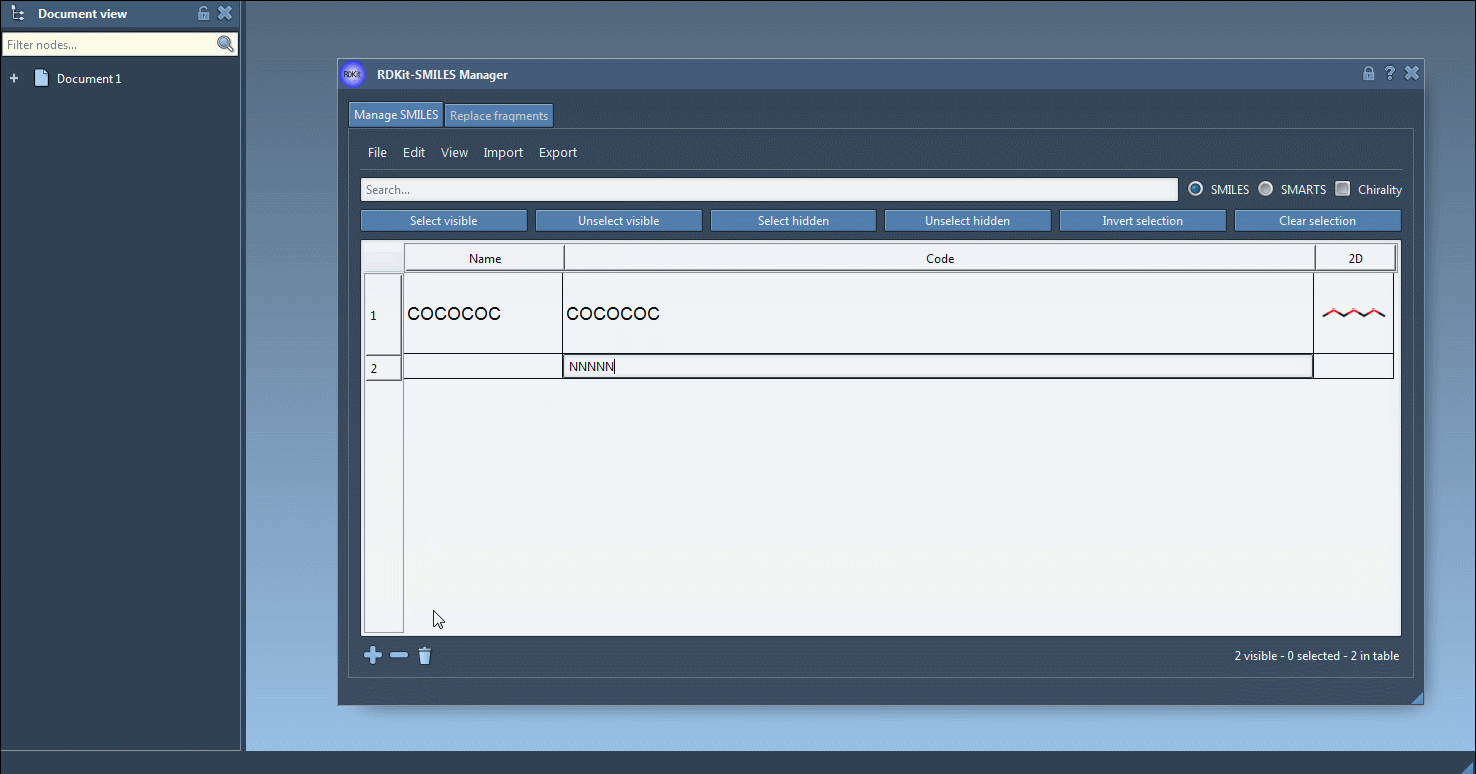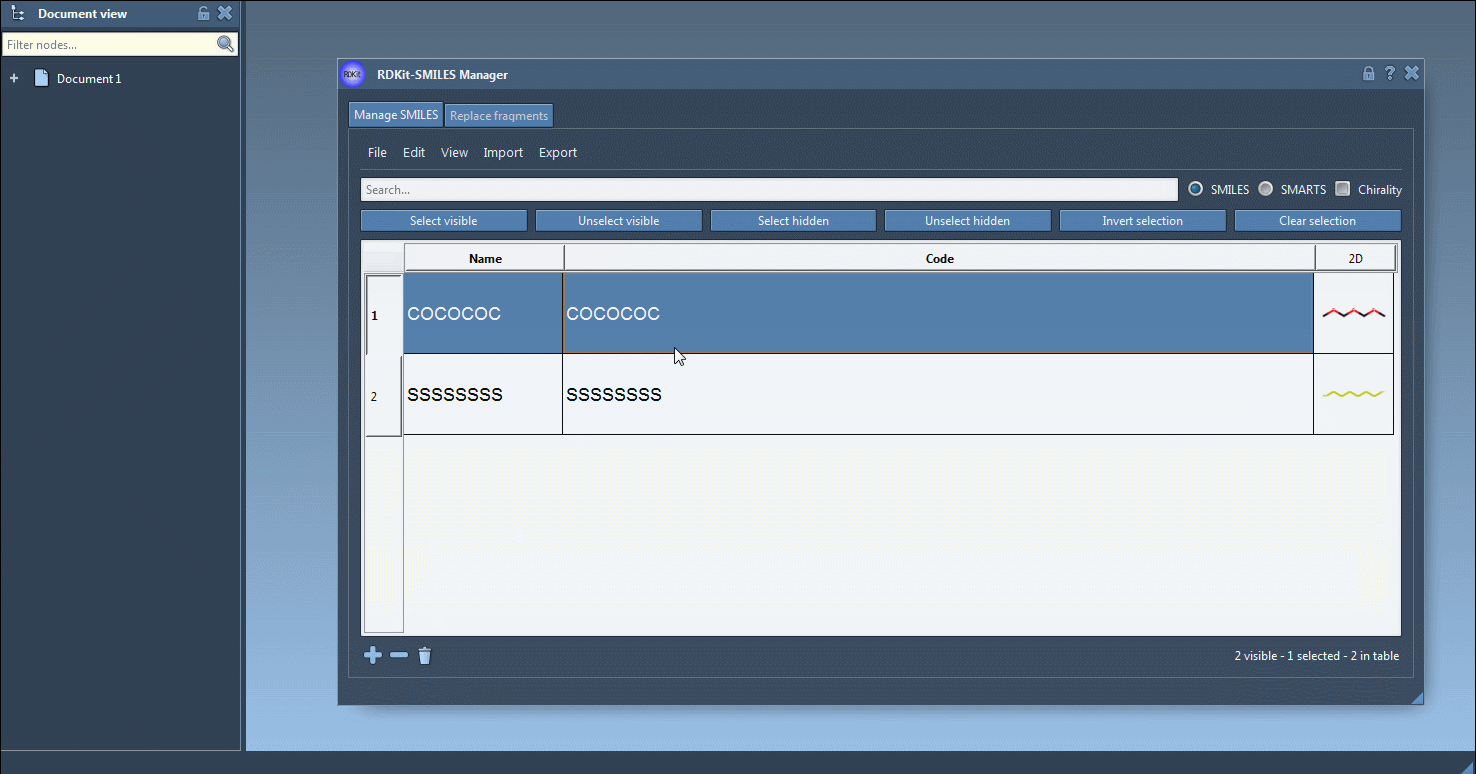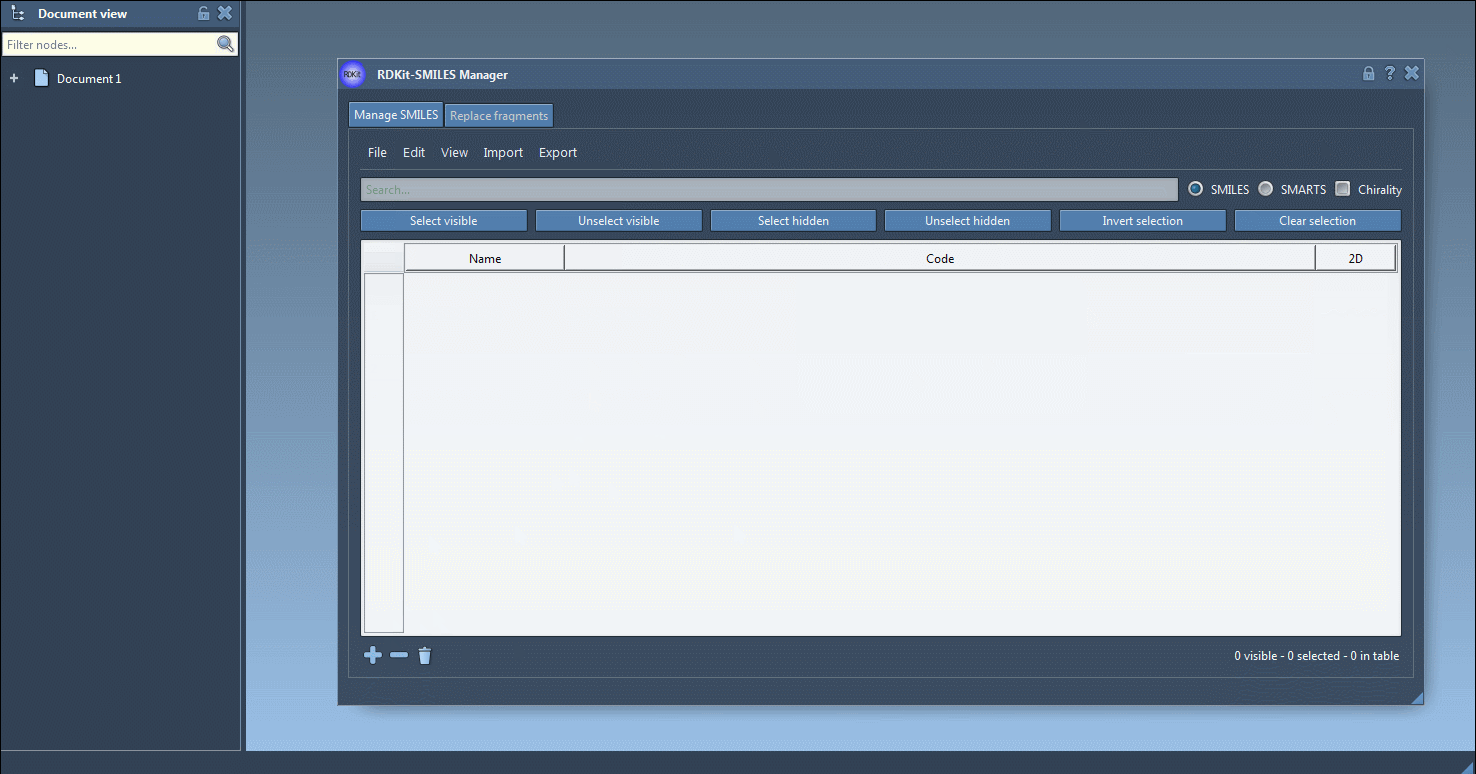
In the Manage SMILES tab, you can load your molecules from a .smi or .txt file. Each SMILES string is displayed in a table alongside its name and automatically generated 2D depiction.
Step 2: Review and Organize
You can add, rename, or edit each SMILES entry manually. The 2D image updates dynamically, giving you visual confirmation of your edits.
Step 3: Generate 3D Structures
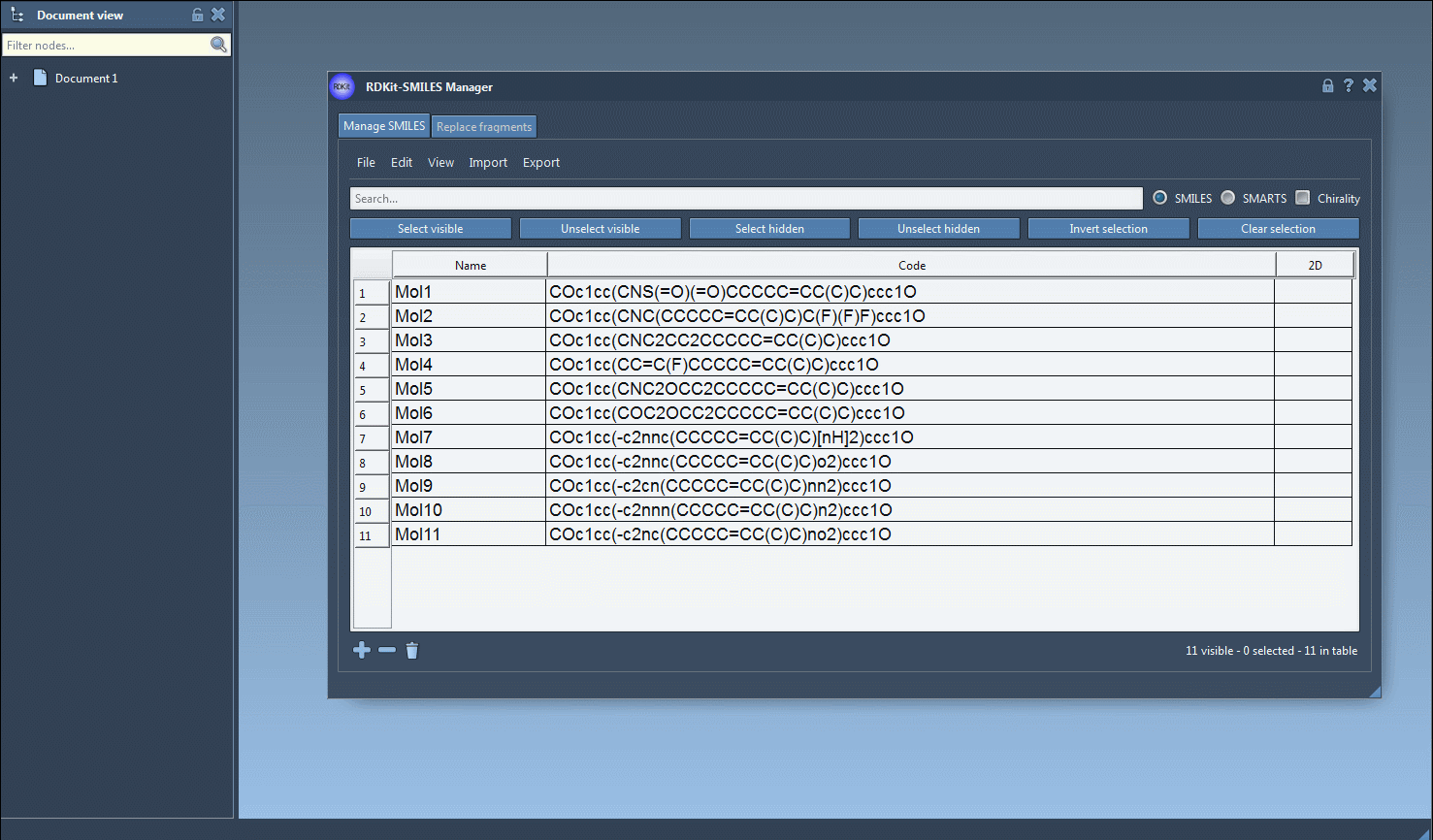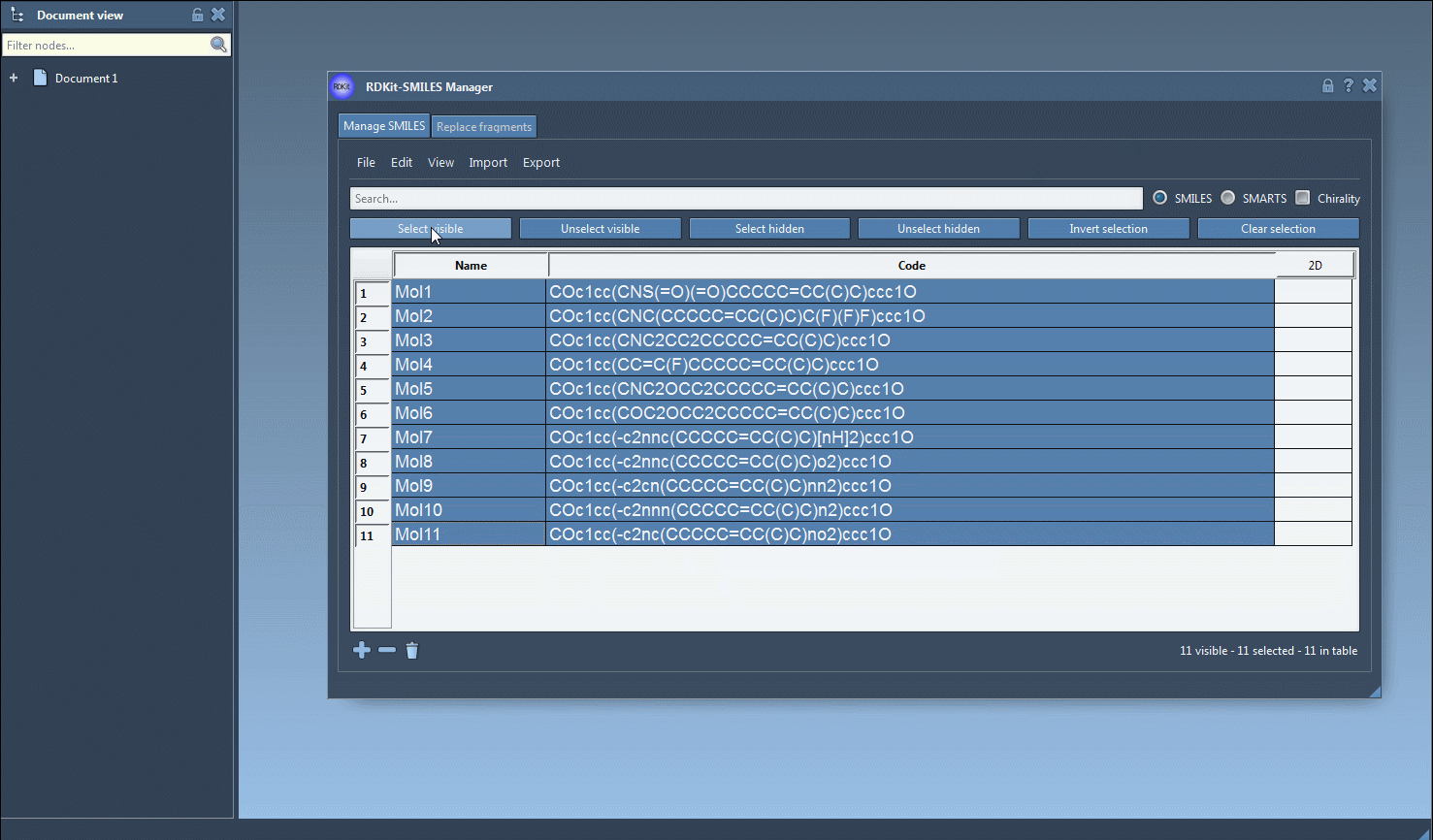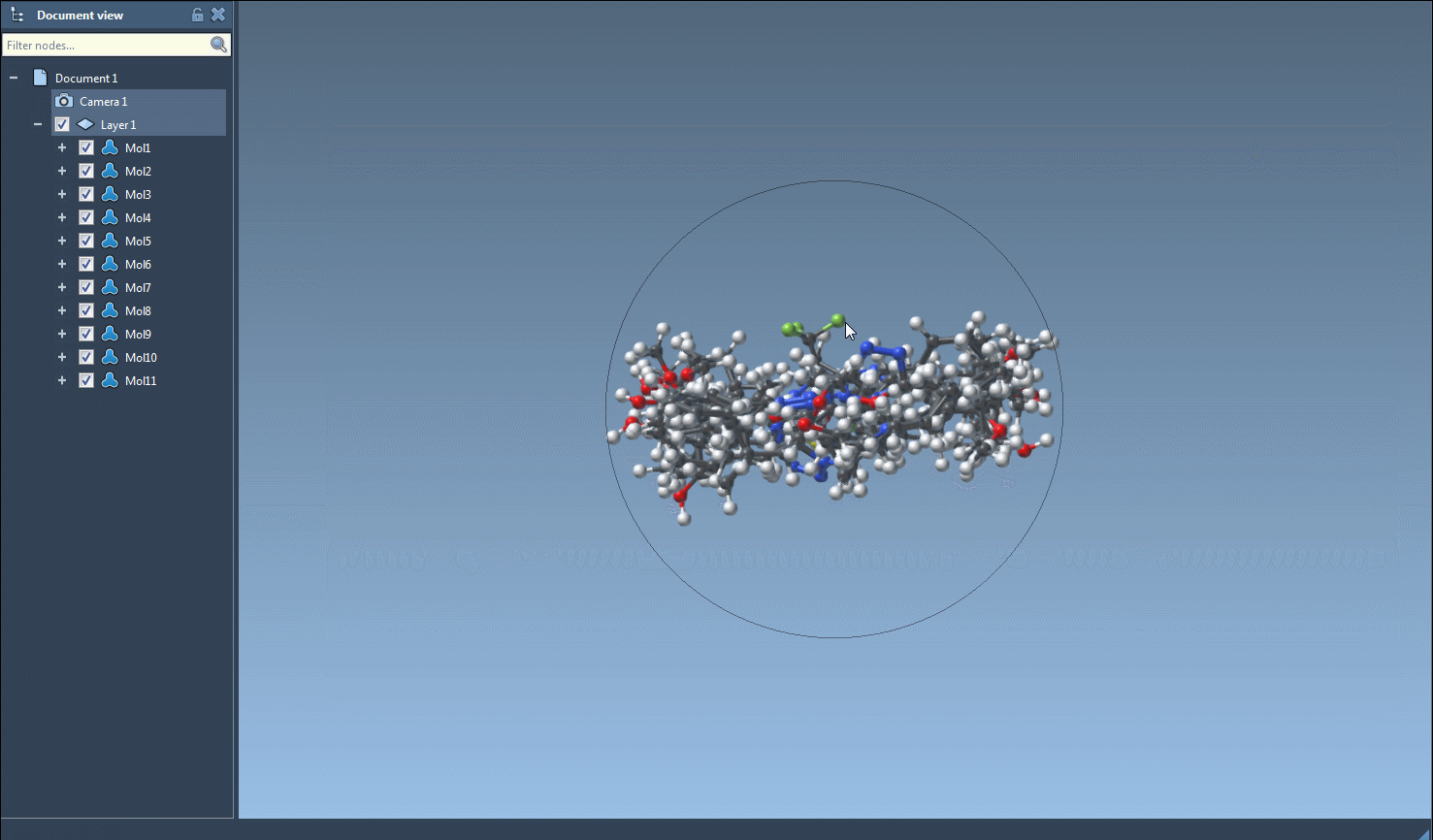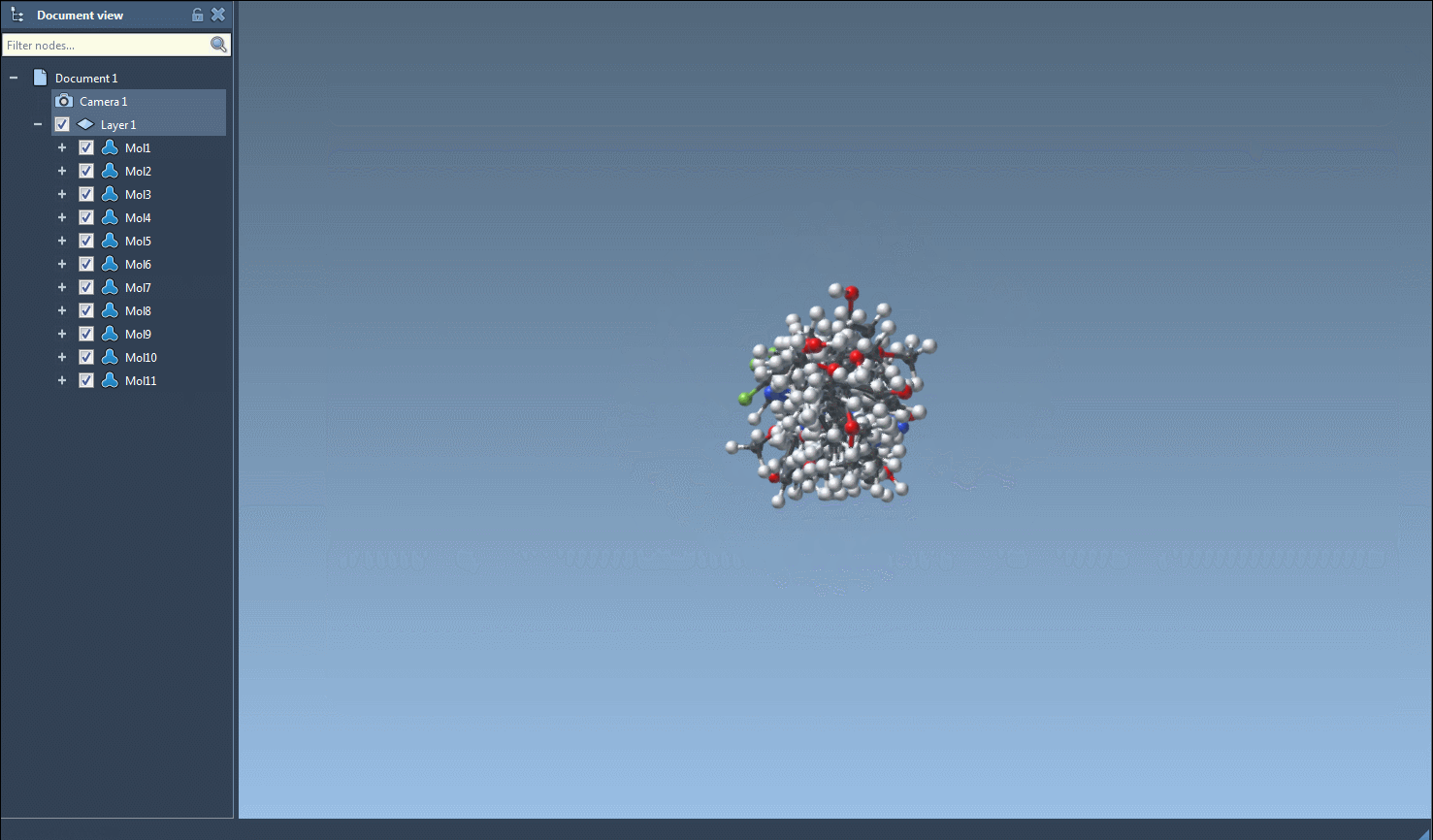
Select one or more SMILES entries in the table. Then, from the Export menu, choose Selected SMILES string to Document. This automatically generates 3D geometries for each selected molecule using RDKit conformation generation and adds them to your current SAMSON document.
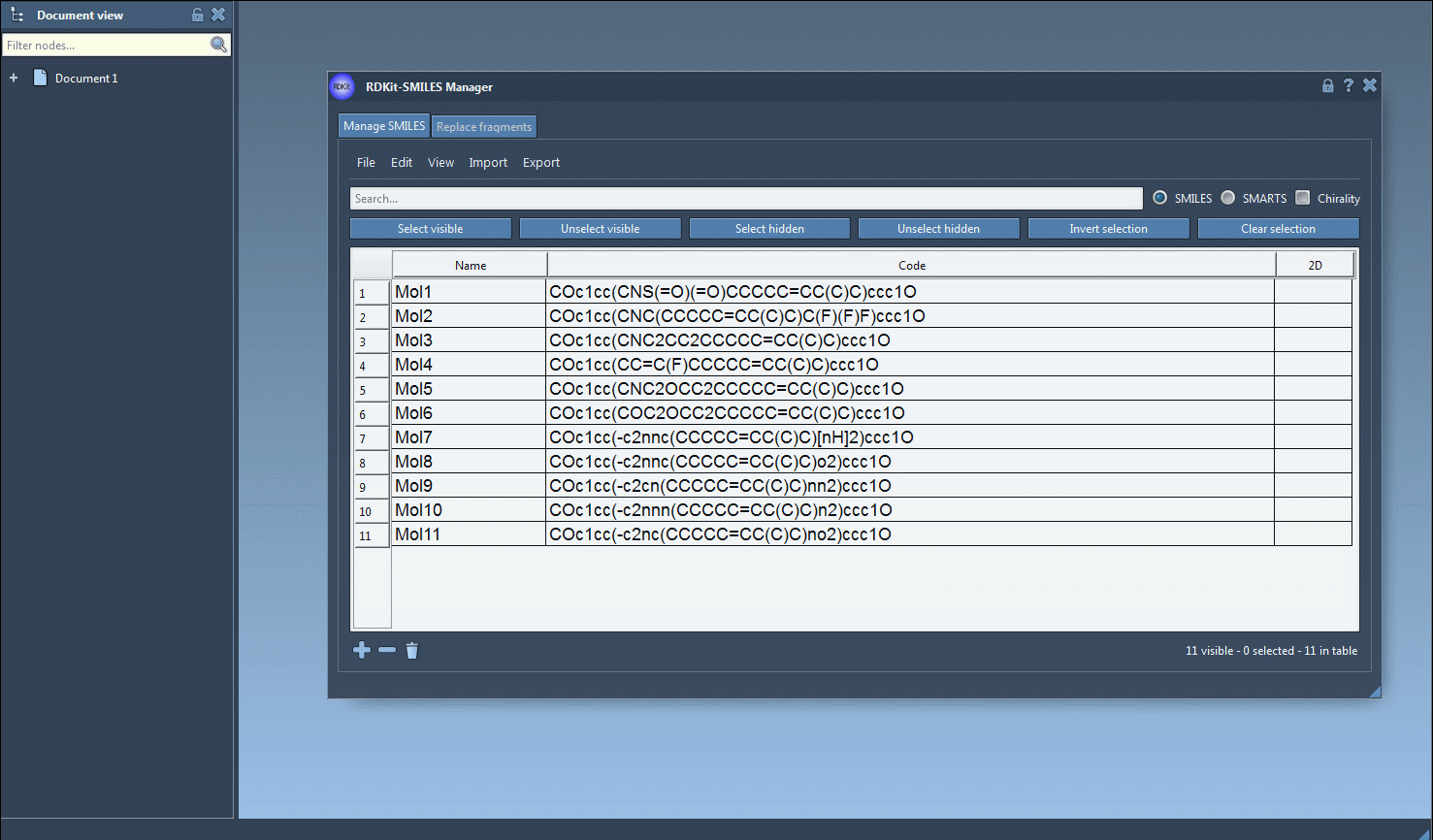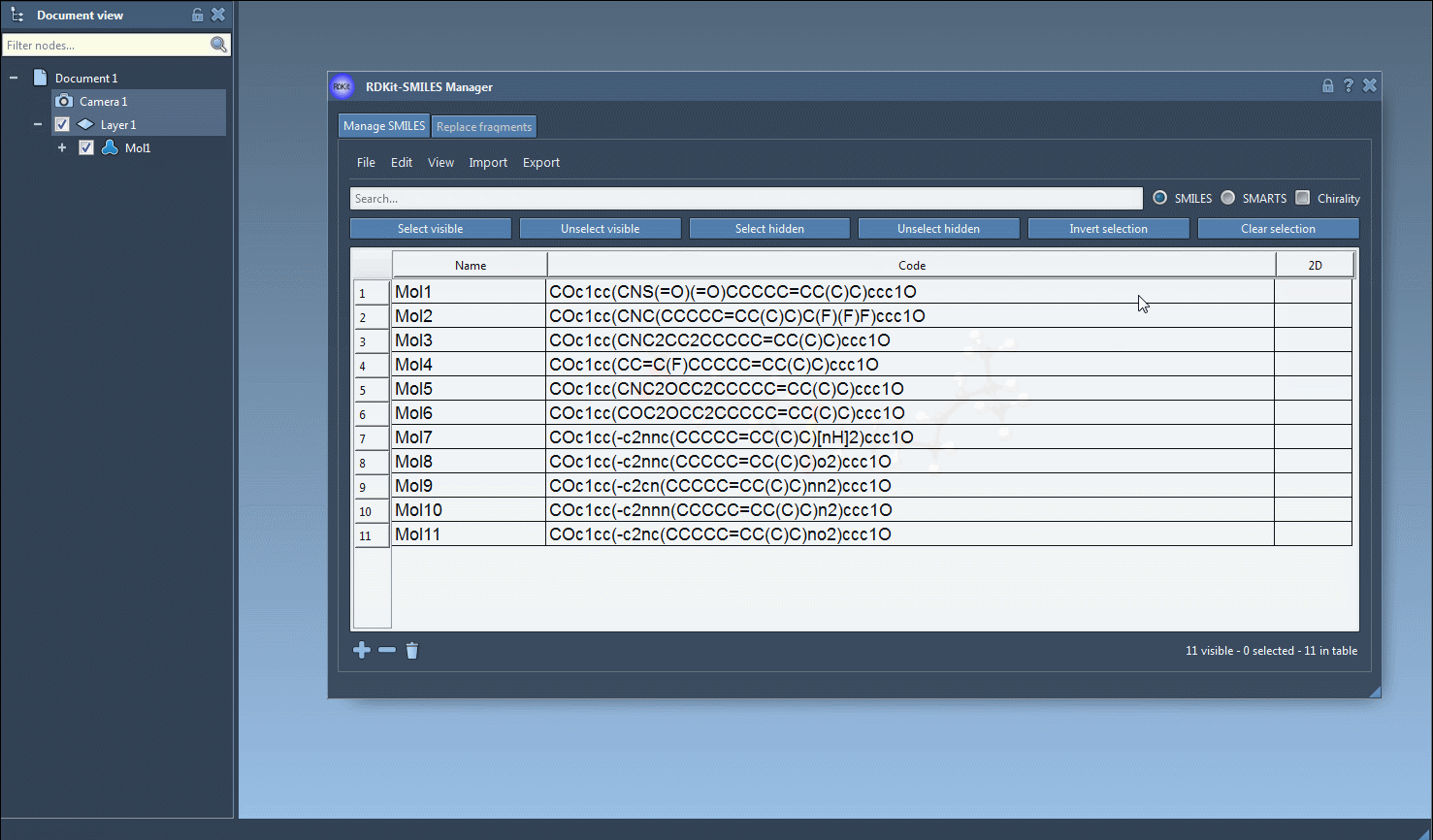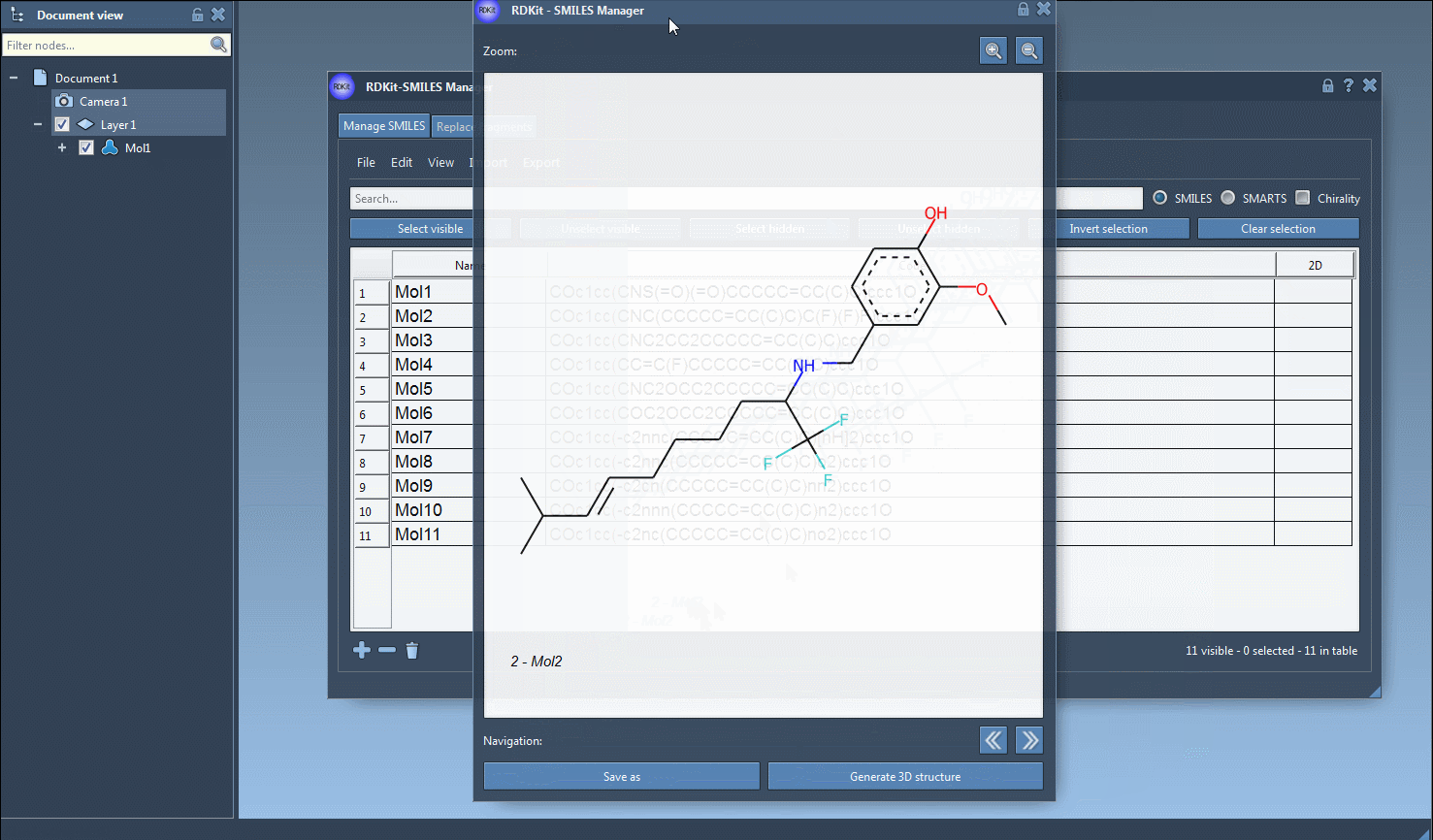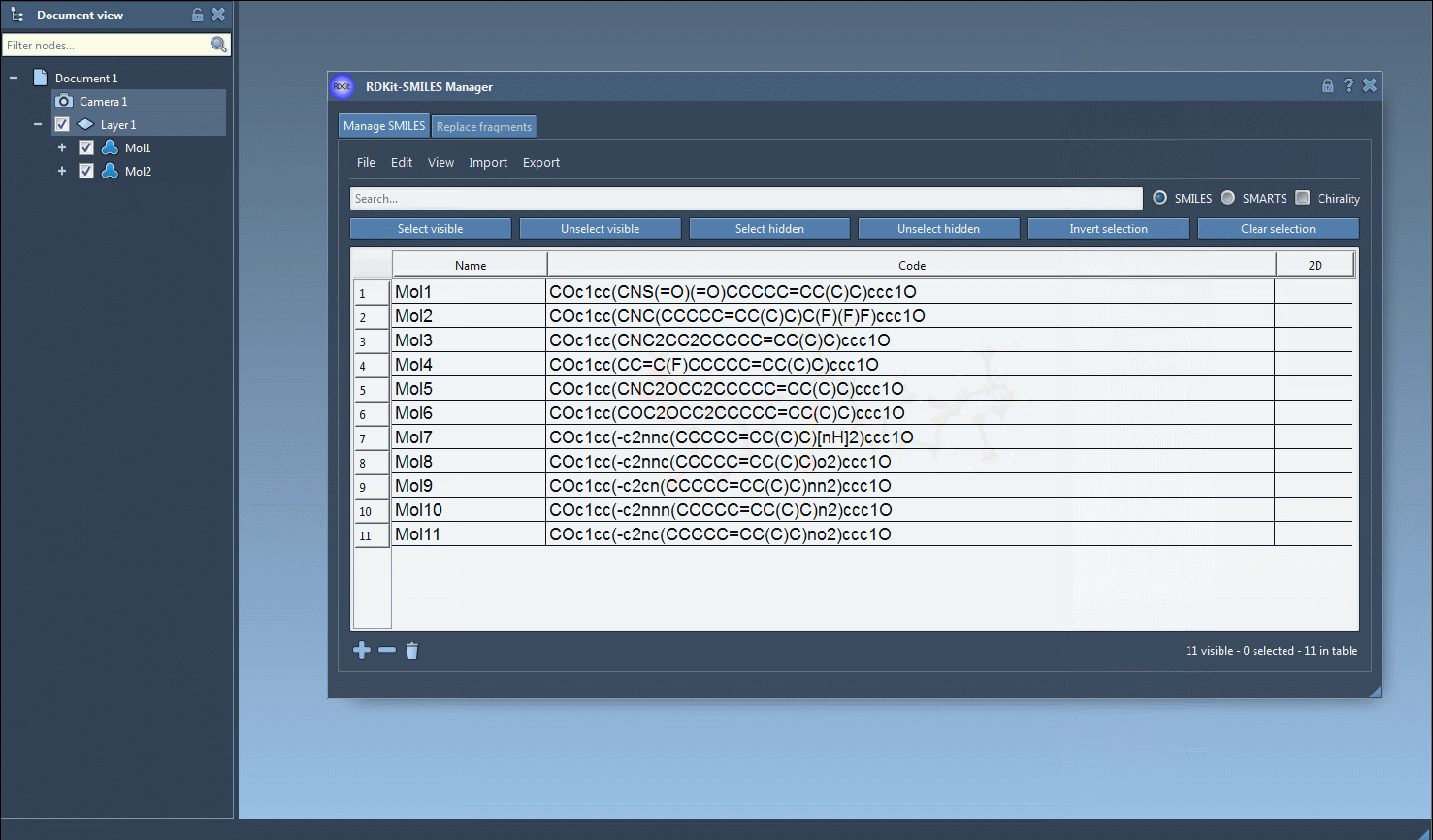
Alternatively, if you prefer to review the molecule before generating 3D coordinates, double-click the 2D depiction to open a larger view. In this view, you can generate the 3D conformation by clicking the Generate 3D structure button.
Step 4: What Happens Next?
Your new 3D molecules are fully integrated into the SAMSON environment. You can immediately visualize them in 3D, run quantum calculations, modify conformations, or perform docking — all within a single interface.
When Should You Use This?
Researchers and modelers often have molecule libraries in SMILES formats and need to quickly transition to 3D for simulation or analysis. SAMSON eliminates the need for scripting or jumping between tools.
This feature is particularly helpful if you:
- Maintain a library of virtual screening candidates in SMILES format
- Want to easily visualize chemical structures with conformations
- Need to prepare input structures for quantum or molecular simulations
To learn more about the SMILES Manager extension and explore additional powerful features such as substructure search and fragment replacement, visit the full documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





