Converting SMILES strings into 3D molecular models can often be a tedious step in molecular modeling pipelines. Whether for QSAR, docking, or structure-based drug design, modelers frequently need to visualize or prepare 3D conformations from simple line notations. While scripting with RDKit or other toolkits is certainly possible, it tends to break the flow for chemists who prefer working visually rather than coding up conversions.
If you’re looking for a quick and reliable way to generate 3D structures from SMILES — without writing code — the Manage SMILES tab of the SMILES Manager extension in SAMSON may be exactly what you need.
This module, built on top of RDKit, offers a graphical interface that lets users import, visualize, and convert SMILES strings into 3D structures in just a few clicks. Here’s how it works:
Drag, drop, convert: How simple 3D generation can be
To get started, users can import a list of SMILES strings from plain-text (.txt) or RDKit-style SMILES (.smi) files. These strings, along with optional names, are shown in a tabular view with real-time 2D renderings generated automatically. This makes it easier to spot structural features and errors.
Once you’re ready to generate 3D structures, just select one or multiple molecules in the table, then click the Export > Selected SMILES string to Document action. This will add the corresponding 3D structures to your active SAMSON document, with geometries derived directly from RDKit.
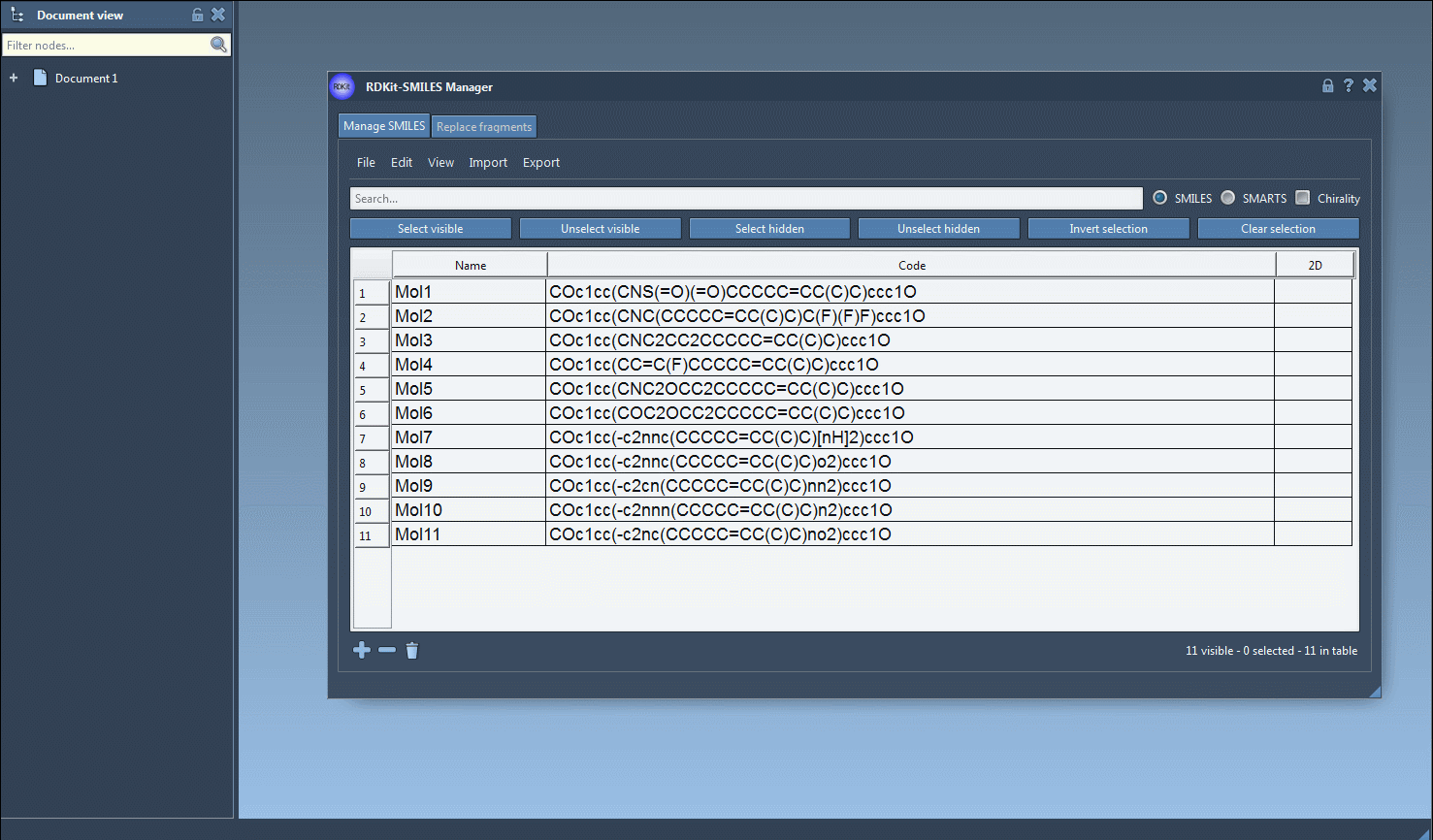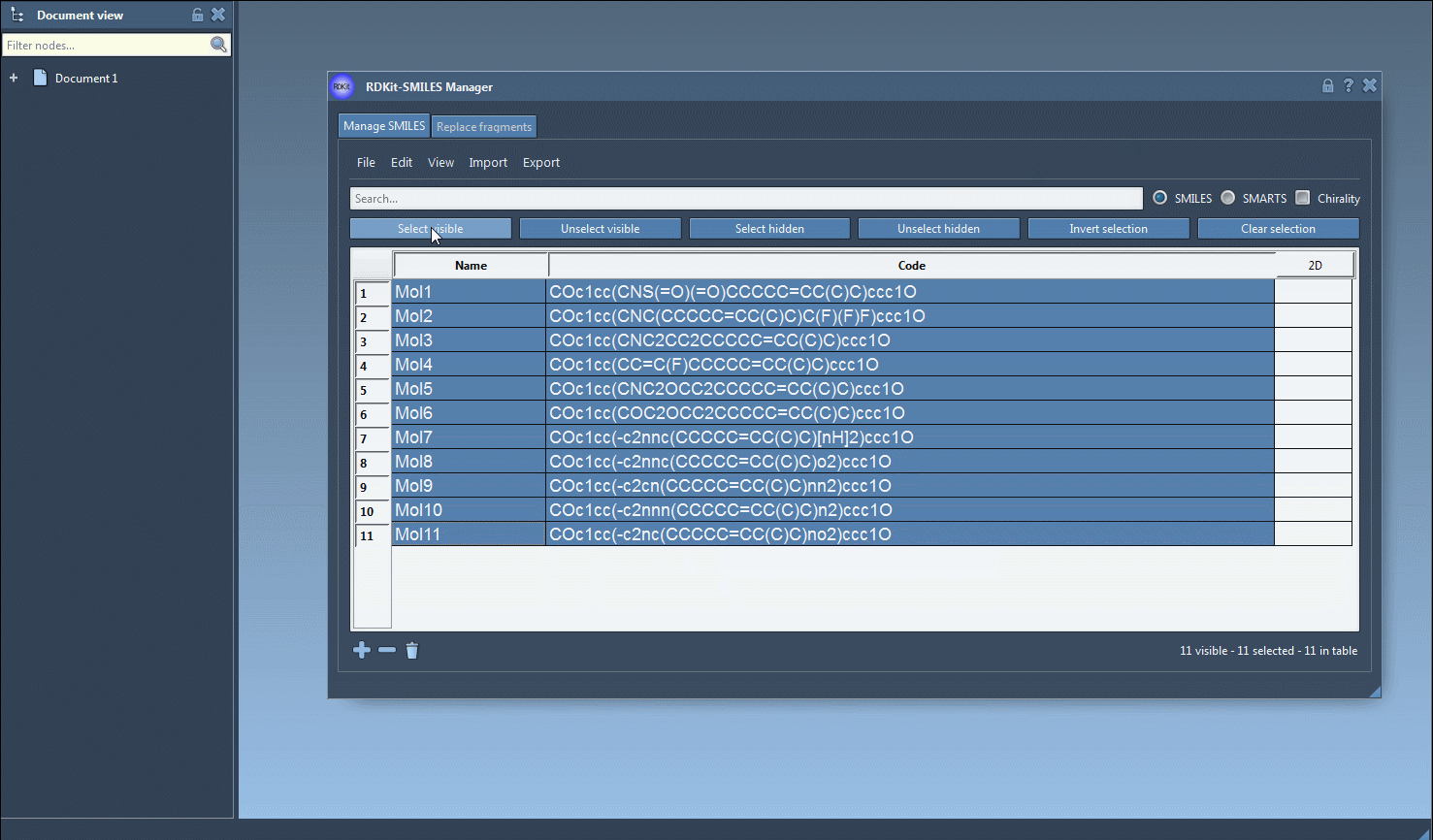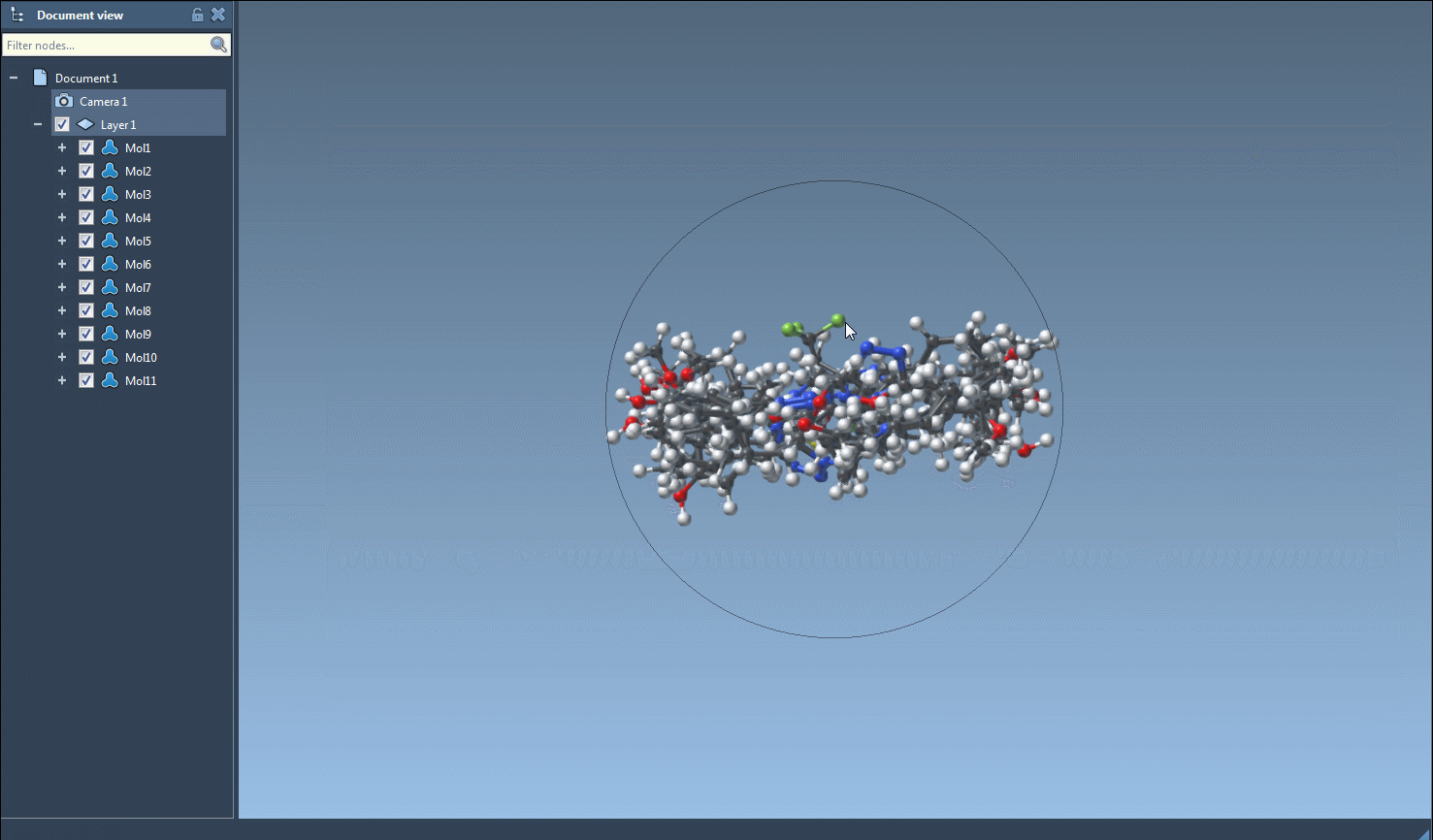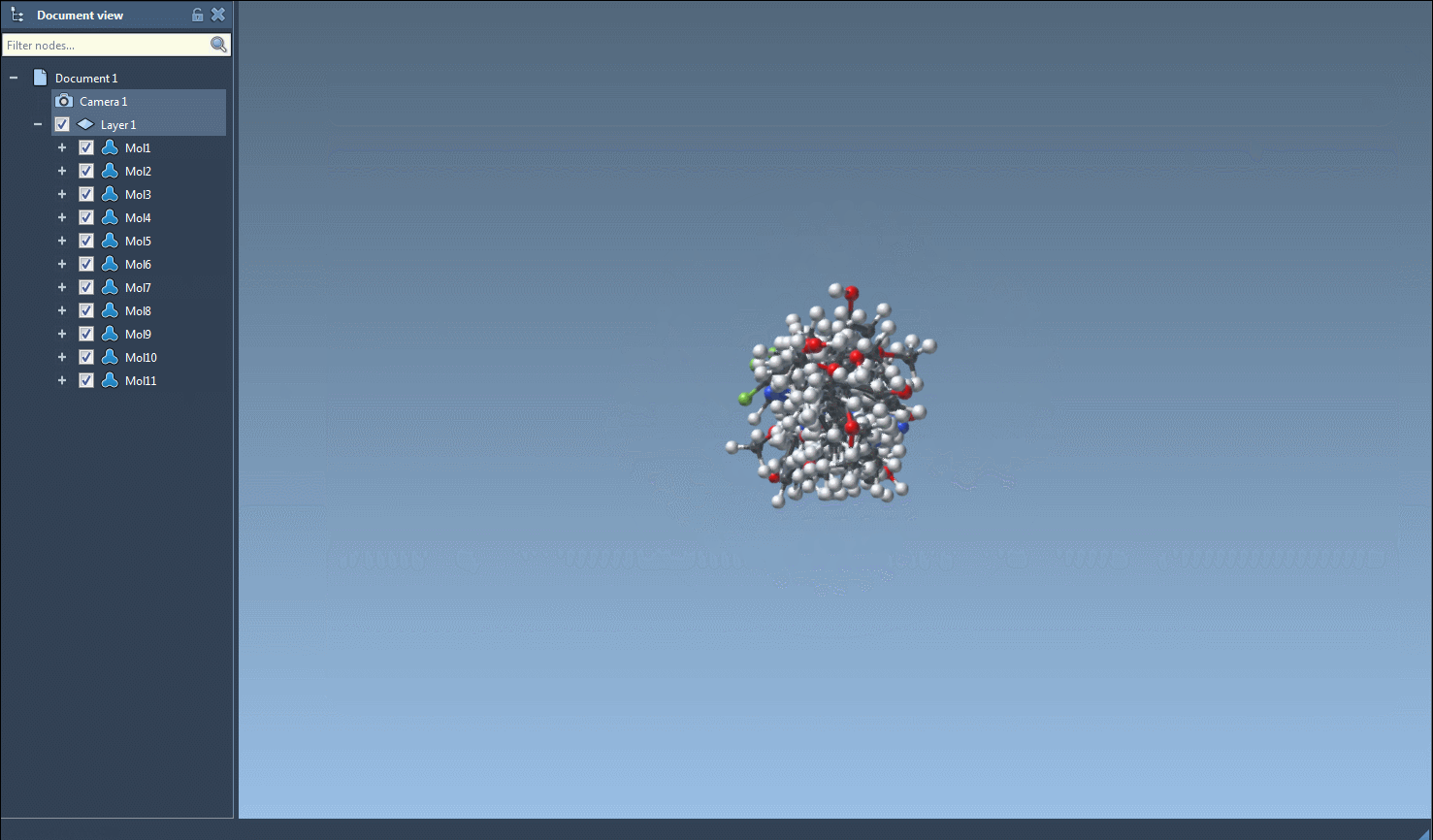
Need a 3D structure for just one molecule? Right-click on a molecule and select Generate 3D structure, or open the molecule’s zoomed-in 2D view and generate from there. It’s flexible and optimized for small workflows as well as library creation.
No more trial-and-error with broken SMILES
Invalid SMILES are automatically flagged and highlighted, and an error image is shown instead of a 2D depiction. This immediate feedback avoids wasted time attempting to generate nonexistent 3D molecules. If you’re working from third-party libraries or manually entered SMILES codes, this validation step is especially helpful.

Bonus: Save and organize visuals
All 2D depictions, including those updated after editing the SMILES, can be saved as PNG or SVG images directly from the interface. That includes a convenient option to export a grid of 2D structures — great for reports or comparing compound libraries.
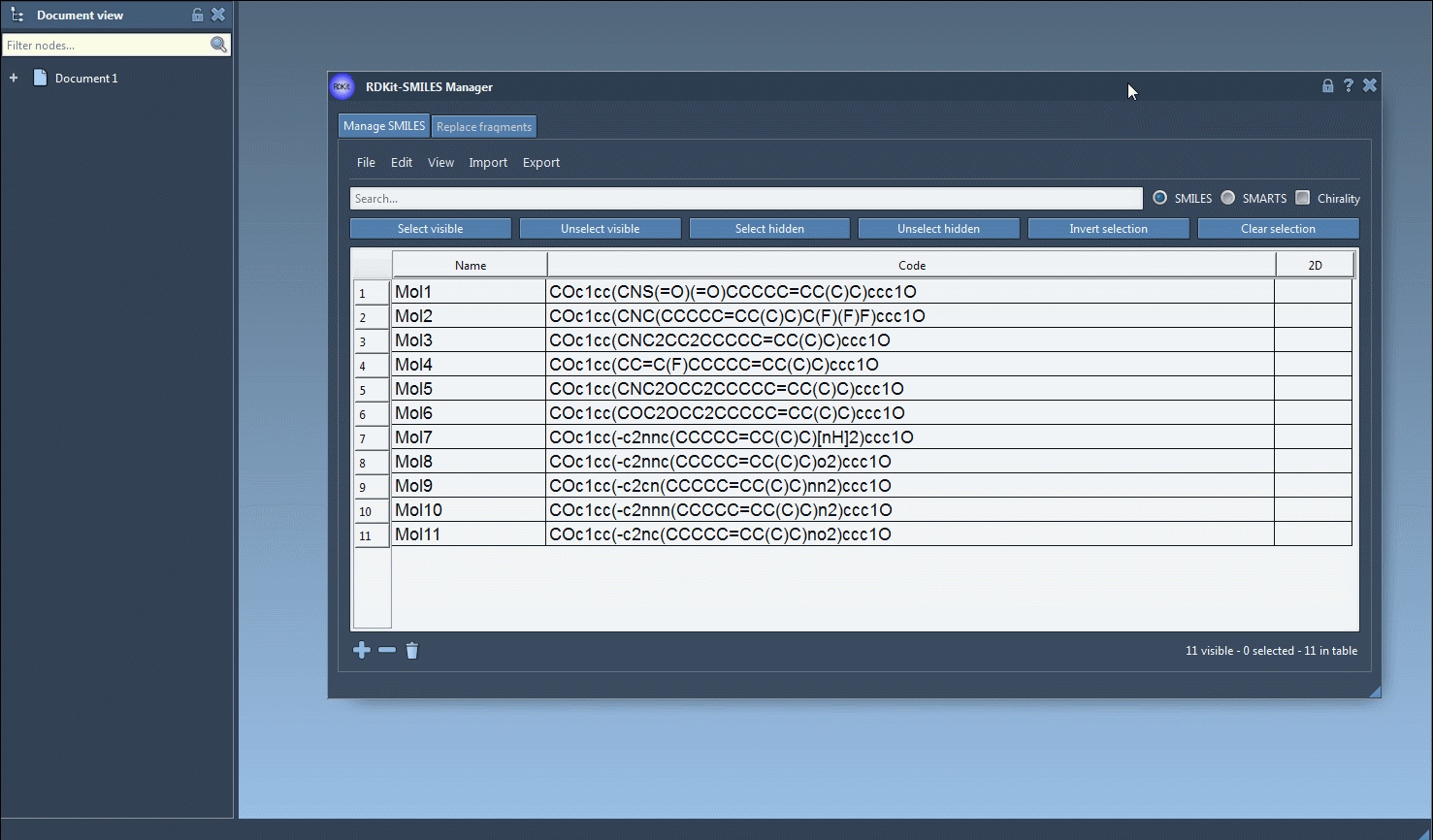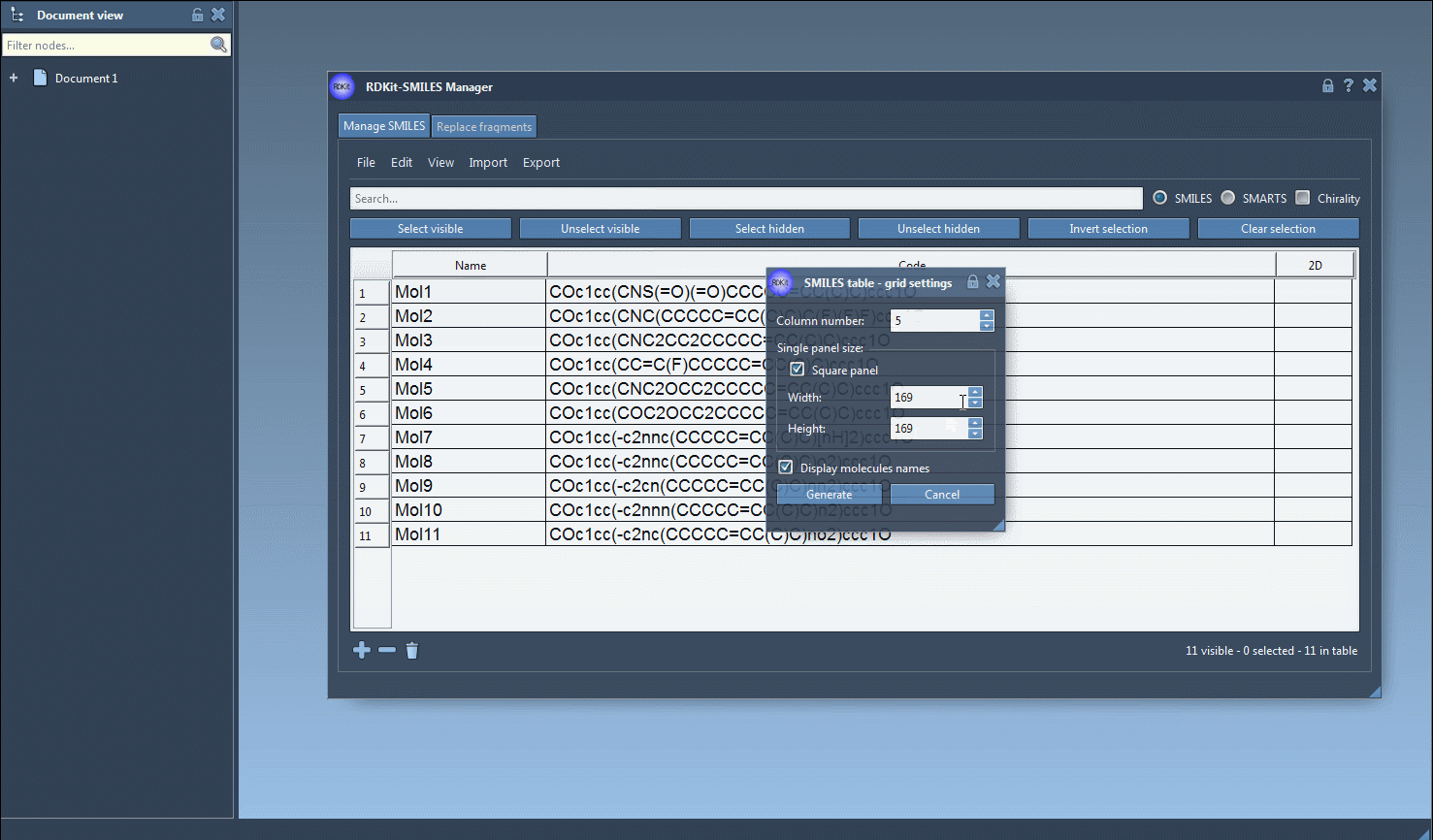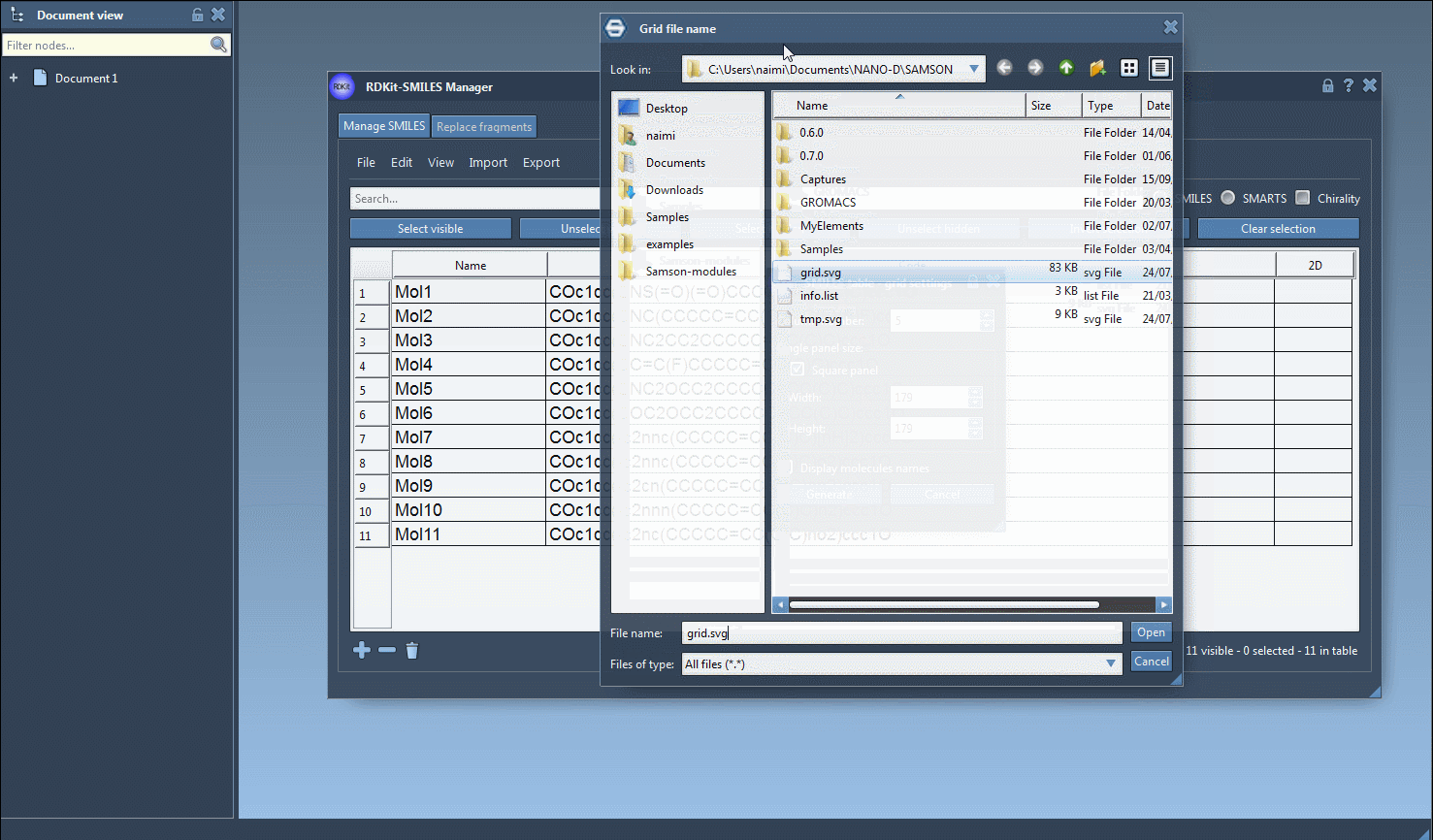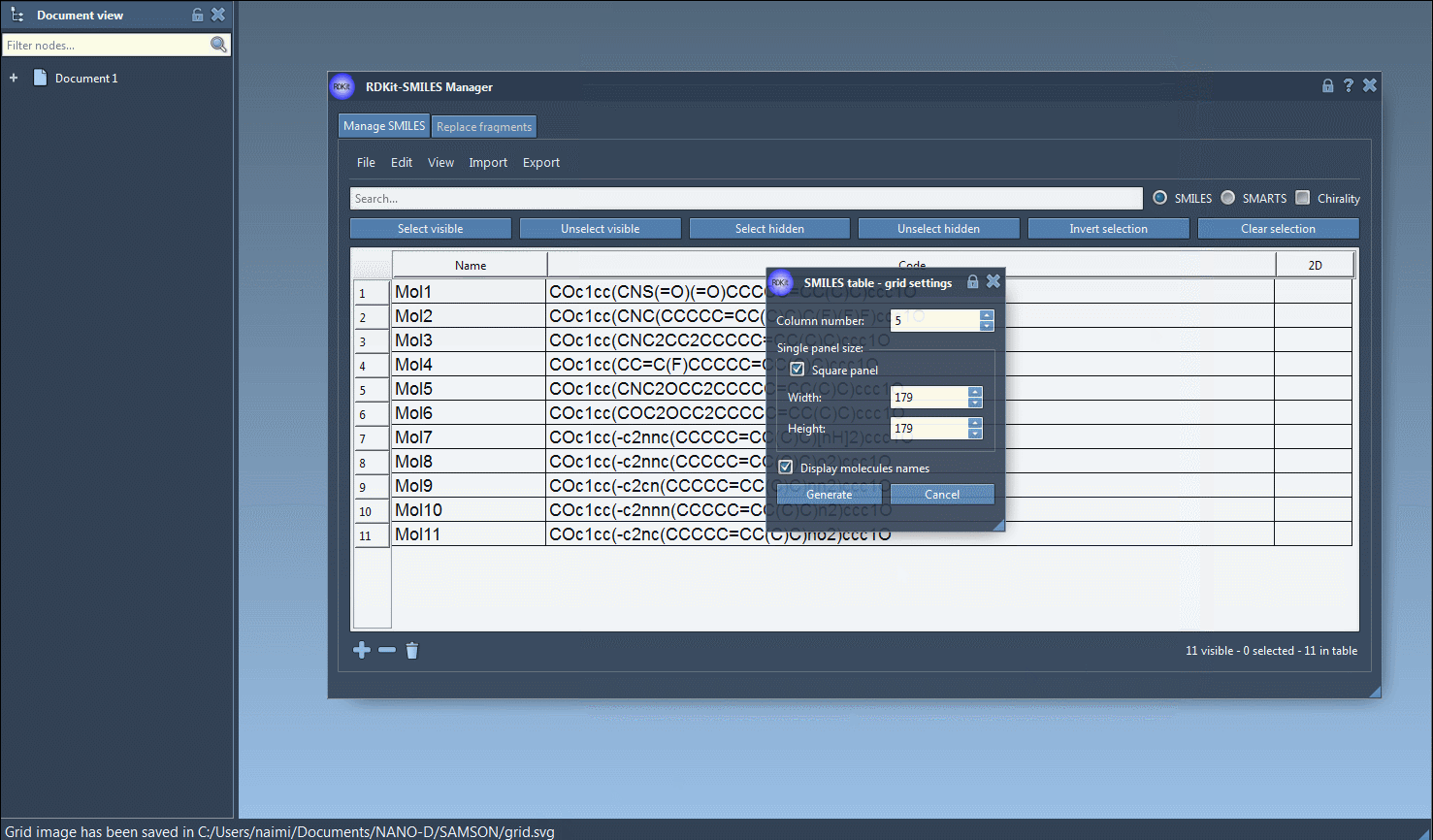
There’s no need to re-type molecular names, either: the software uses the assigned name or defaults to the SMILES string if none is provided. Everything is editable in place.
Why it matters
For modelers who frequently receive compound lists in SMILES format from collaborators or databases, being able to generate clean 3D structures on the fly is a real time-saver. It simplifies workflows when evaluating candidates, preparing input for simulation tools, or designing libraries for further exploration.
This functionality lowers the barrier to producing consistent, valid molecular geometries without having to invoke Python scripts or pre-process files manually. If you’re visual and like to work with tools that offer feedback as you go, it’s a very useful capability.
Learn more about this and other SMILES Manager features in the full documentation: SMILES Manager User Guide.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON here.





