If you’ve ever opened a PDB file and been disappointed to find only part of the structure—just one chain or a fragment of a protein complex—you’re not alone. Many researchers assume PDB files always include the full biological assembly. Instead, these files often contain only the asymmetric unit, lacking the symmetric partners that form the complete quaternary structure.
This can be a roadblock in workflows involving protein-protein interfaces, oligomeric assembly analysis, or nanostructure design. If you’re manually reconstructing missing partners or turning to external tools before returning to your workspace, there’s an easier way: the Symmetry Mate Editor in SAMSON.
What Are Symmetry Mates and Why Do They Matter?
Symmetry mates are structural replicas generated from transformation data included in the PDB file—either in the CRYST1 record (for crystal symmetries) or BIOMT records (for biological assemblies). Viewing these symmetric partners helps model realistic complexes, predict interaction surfaces, and understand molecular function in context.
Using symmetry mates is a routine part of modeling, yet many tools make the process tedious or opaque. Integration with SAMSON simplifies the process by making replication modular, visual, and reversible.
Instantly Reconstruct Full Assemblies in SAMSON
Once you install the Symmetry Mate Editor extension (free for non-commercial use), activate it with:
- Shift + E and search for “Symmetry Mate Editor”, or
- Use the left-side menu: … > General > Symmetry Mate Editor
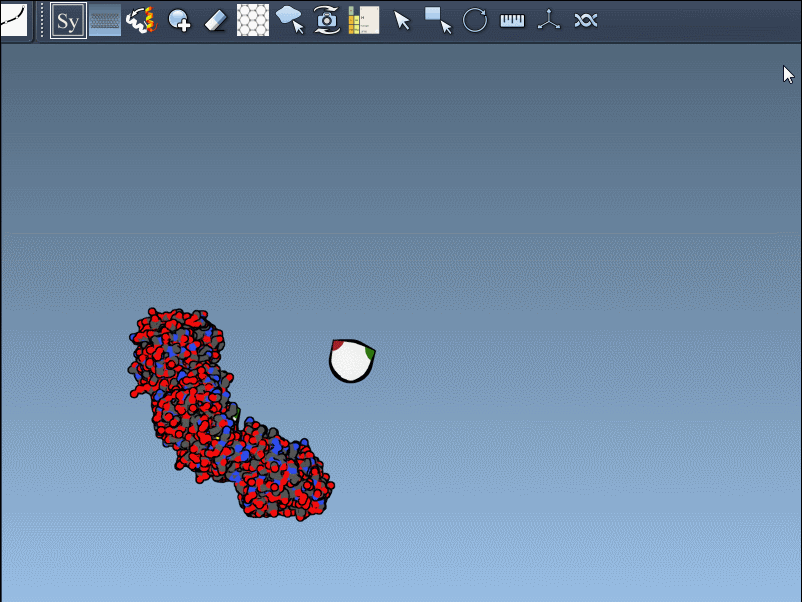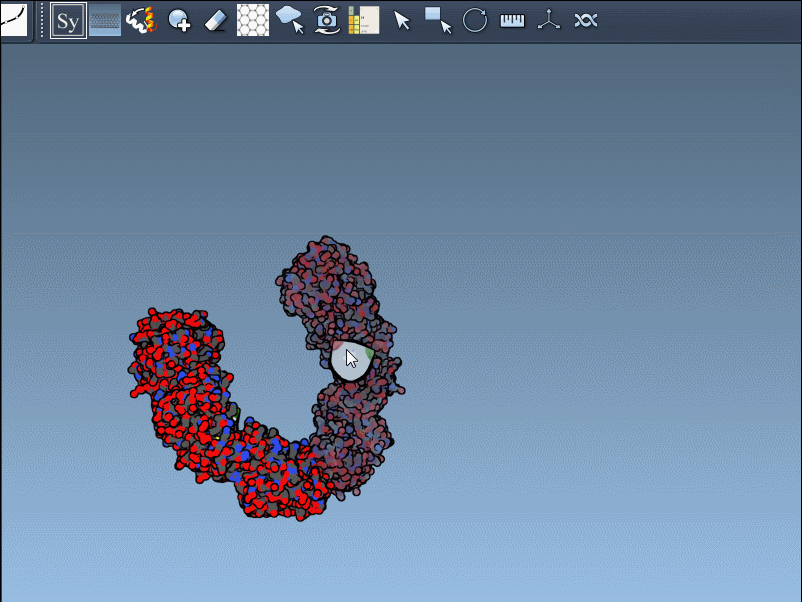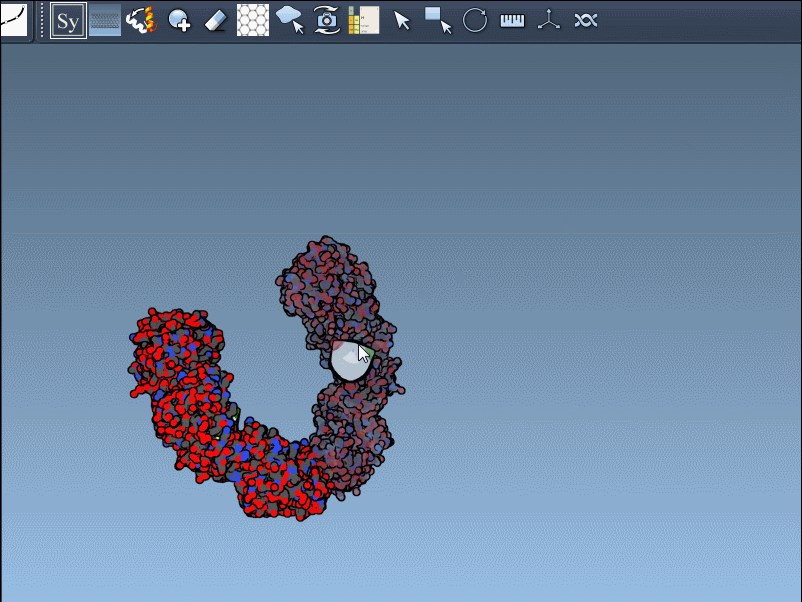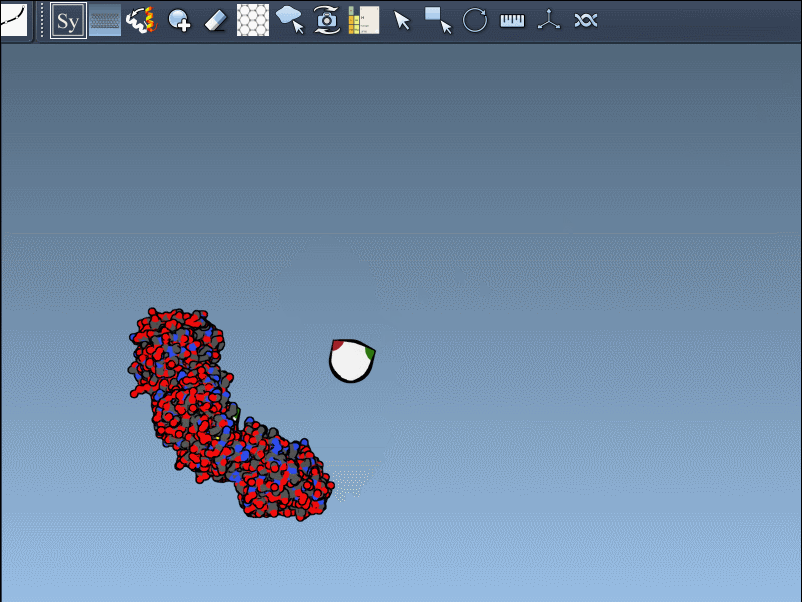
This brings up a 3D interface with control nodes around your protein model. These nodes represent available symmetry transformations.
Preview Before You Generate
Hover over a control node to preview a symmetry mate replica in real time. This interactive feature helps identify useful symmetries without unnecessary clutter:
Click a node to generate and place the replica permanently—or hold Ctrl/Cmd while clicking to generate all available mates in one step:
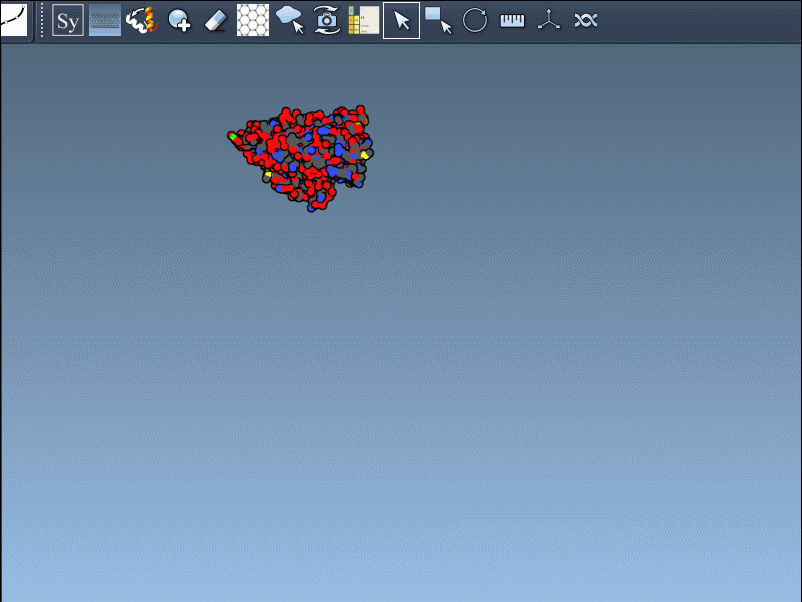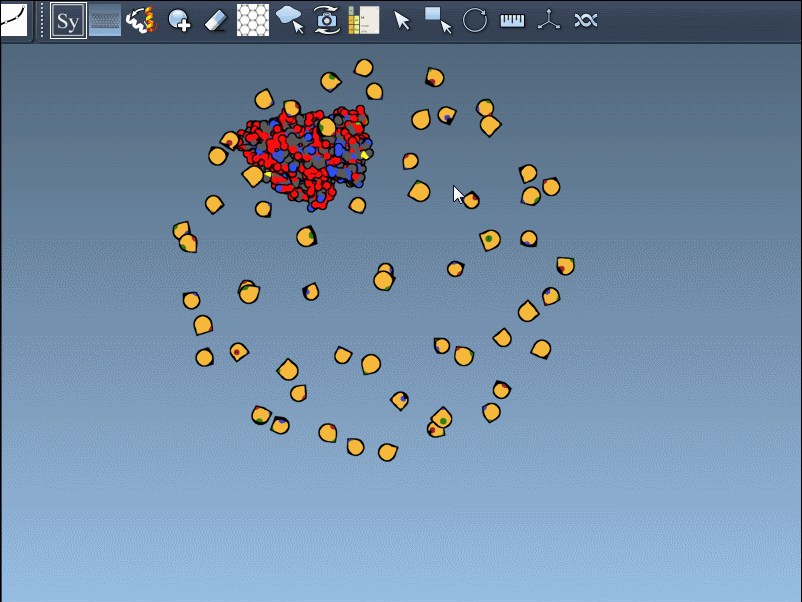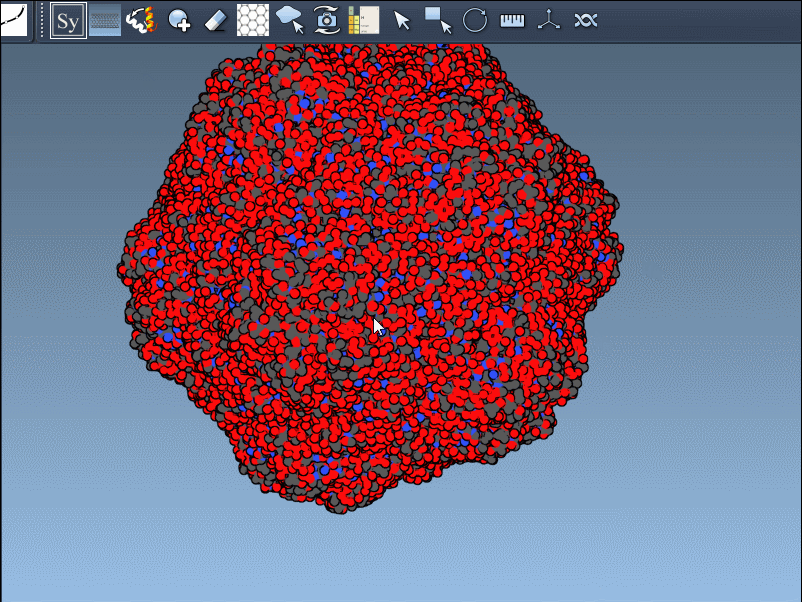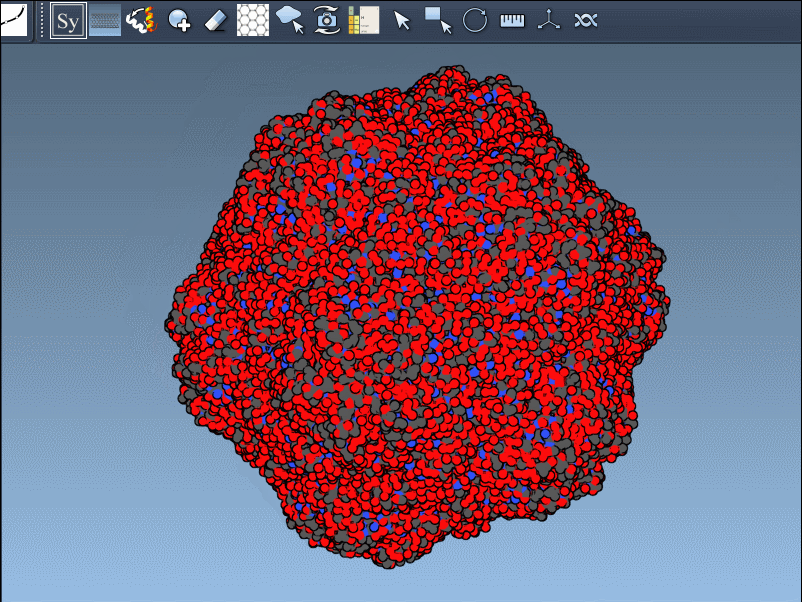
Switch Between Crystal and Biological Assembly Symmetries
The PDB format may contain both CRYST1 and BIOMT data. SAMSON distinguishes these with different widget colors:
| Type | Source | Color |
|---|---|---|
| CRYST1 | Crystal lattice symmetry | White |
| BIOMT | Biological assembly | Yellow |
You can switch between these symmetries to compare how the protein is arranged in crystals versus its functional biological form:
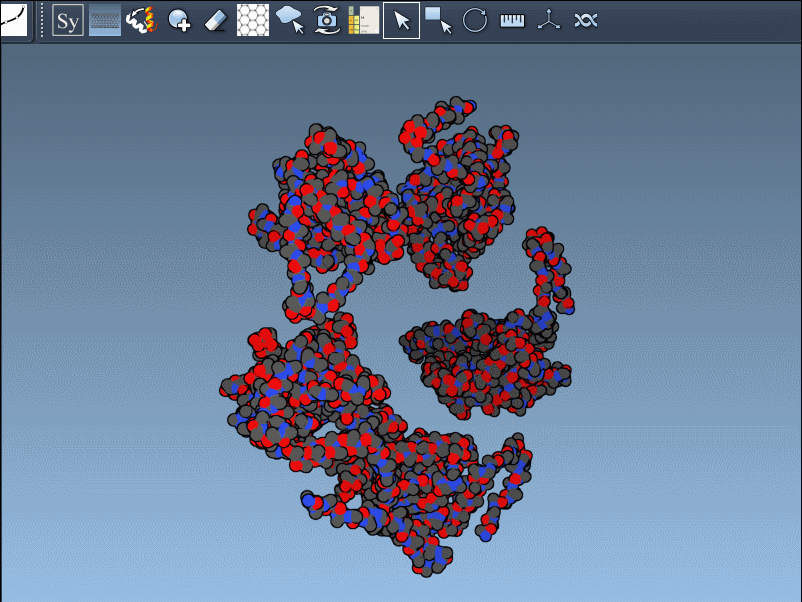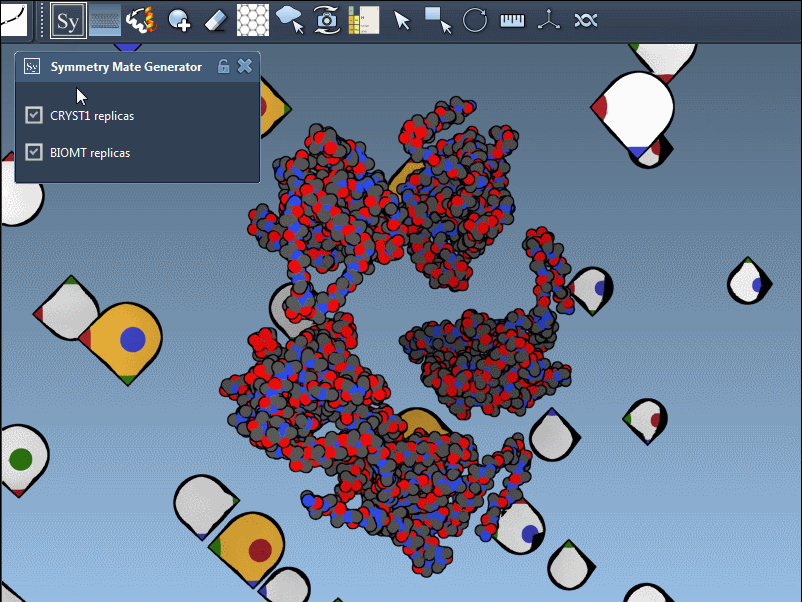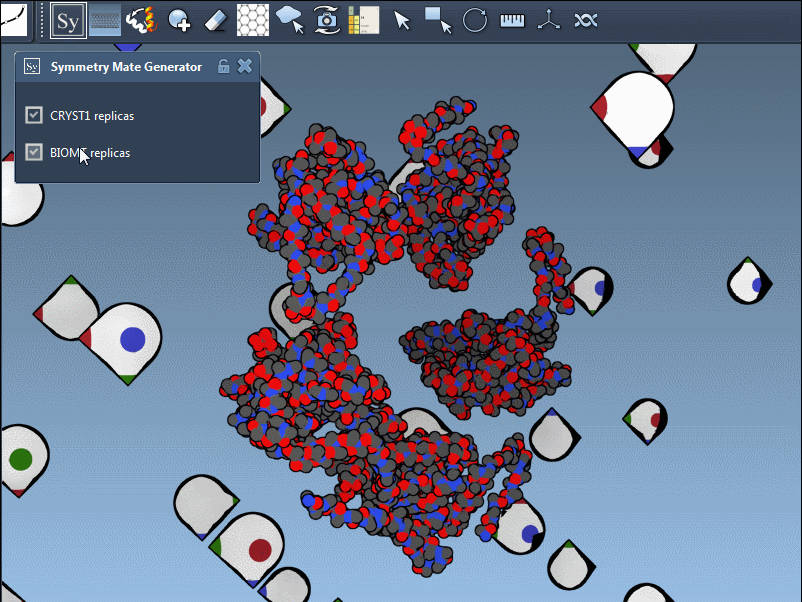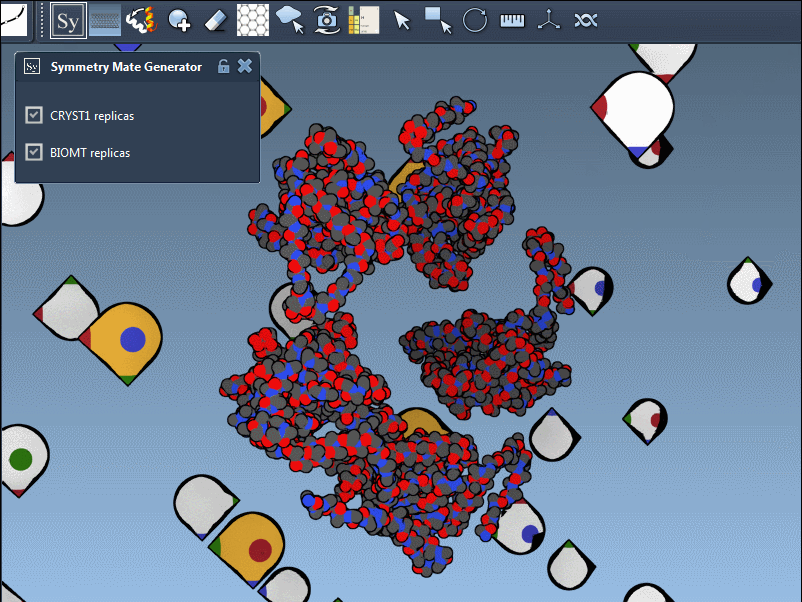
Use Cases for Molecular Modelers
- Evaluate and model protein-protein interactions at full interface surfaces.
- Design symmetric nanostructures based on oligomeric templates.
- Simulate dynamics of entire assemblies instead of isolated chains.
Final Tip
You can always use Ctrl/Cmd + Z to undo generated replicas if needed—experiment freely!
To explore these features in depth, visit the original documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON here.





