When designing complex polymer structures, one common challenge for molecular modelers is efficiently defining and managing sequences of monomer units. Particularly in synthetic or biopolymer design, having both precision and flexibility in how monomers are arranged is crucial for building realistic molecular scaffolds. If you’ve ever struggled to manage variations, repeat units, or custom connection rules between monomers, SAMSON’s Polymer Builder provides a practical solution.
Let’s focus on a highly useful but sometimes overlooked feature: the ability to create and register monomer sequences. This capability helps bring order to what could otherwise become a very manual and error-prone process, especially for large or repetitive polymers.
Registering Custom Sequences: Why It Matters
After registering individual monomers in SAMSON’s Polymer Builder, you can combine them into custom sequences — a key step when working with polymers that follow a particular motif or structural pattern. Instead of manually repeating elemental monomer identifiers for each new chain, you can:
- Group registered monomers into reusable sequences
- Assign a name to each sequence for better organization
- Preview and validate sequences before generation
- Specify bond types within sequences: single, double (using ‘=’), or triple (‘#’)
For example:
ABBAuses monomers A and B in a standard configuration.A=B: A double bond connects monomers A and B.A#B: A triple bond is enforced between monomers A and B.
This makes it easier to model polymers like conjugated materials, block copolymers, or increasingly popular sequence-controlled polymers—all essential structures for materials science and bioengineering research.
Visual Feedback Built In
One of the practical strengths of the Polymer Builder is immediate feedback. After entering your new sequence with monomer IDs, the app validates your input. Syntax errors (like referencing unregistered monomers or incorrect bond syntax) are immediately shown in the status column.
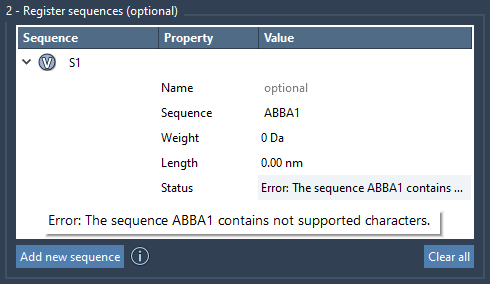
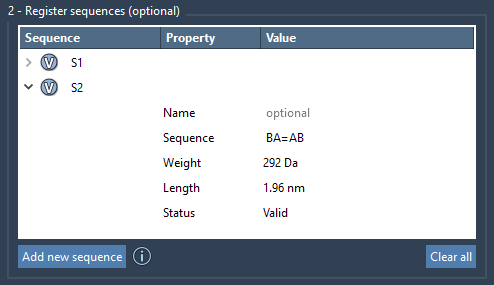
When a sequence is valid, it will be labeled and stored with a unique identifier, like S1, S2, and so on, which you can later reuse while generating polymers.
Add, Modify, or Remove Sequences
The workflow is streamlined:
- Click Add new sequence
- Enter the desired pattern (e.g.,
ABBAorA=B#C) - Optionally specify a name for grouping purposes
You can edit a sequence’s name or composition directly in the table, preview the result by clicking the associated V button (highlight monomers visually), and delete or clear sequences if needed.
Why Use this Workflow?
This sequencing approach reduces redundancy, minimizes the chance of structural mistakes, and prepares your model for downstream tasks—like energy minimization or export to GROMACS or LAMMPS. It’s also adaptable enough to support polymers with mixed bond types, modular blocks, and alternative branching options.
If you’re building molecules for simulations, electronic properties research, or functionalized materials, sequence control provides an essential layer of structural clarity and experiment reproducibility.
To explore this feature and more, visit the full documentation here: SAMSON Polymer Builder Documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON here.





