When setting up molecular dynamics simulations, embedding a protein within a lipid membrane can be time-consuming and error-prone. Whether you’re studying membrane proteins, simulating transmembrane transport, or modeling realistic biological environments, creating these lipid environments from scratch often requires multiple tools and extensive manual setup.
The Molecular Box Builder extension in SAMSON streamlines this process by allowing users to easily build molecular boxes filled with customized arrangements of lipids around proteins. In this post, we focus on creating a single lipid layer around a protein—quickly and reproducibly—without relying on external membrane builders or scripting.
Aligning Your Protein
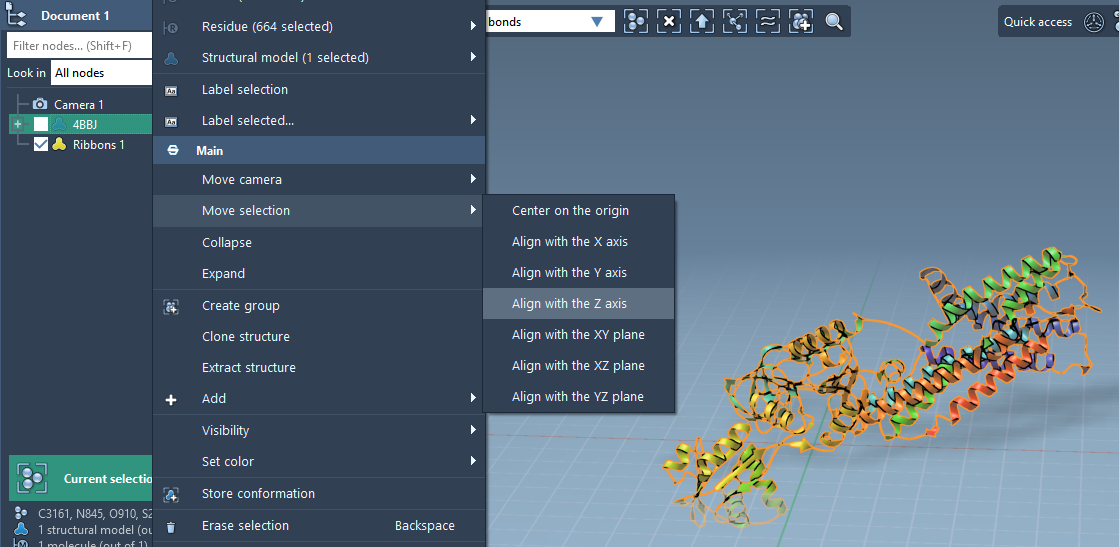
Accurate alignment is key to generating a functional membrane-protein system. You can interactively reorient the protein within SAMSON:
- In the Document view, right-click your protein model.
- Choose Move selection > Align with Z axis. This ensures the membrane will form perpendicular to Z, resembling its orientation in a cellular environment.
- Then choose Move selection > Center on the origin to place the protein at the center of the scene.
Selecting and Aligning the Lipid
- Load or import your desired lipid molecule (e.g., POPC).
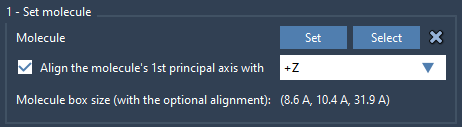
- Select the lipid structure within the Viewport or Document view and click Set in the Molecular Box Builder app.
- Align its principal axis to the
+Zdirection. This ensures lipid tails orient uniformly.
Defining the Box Around the Protein
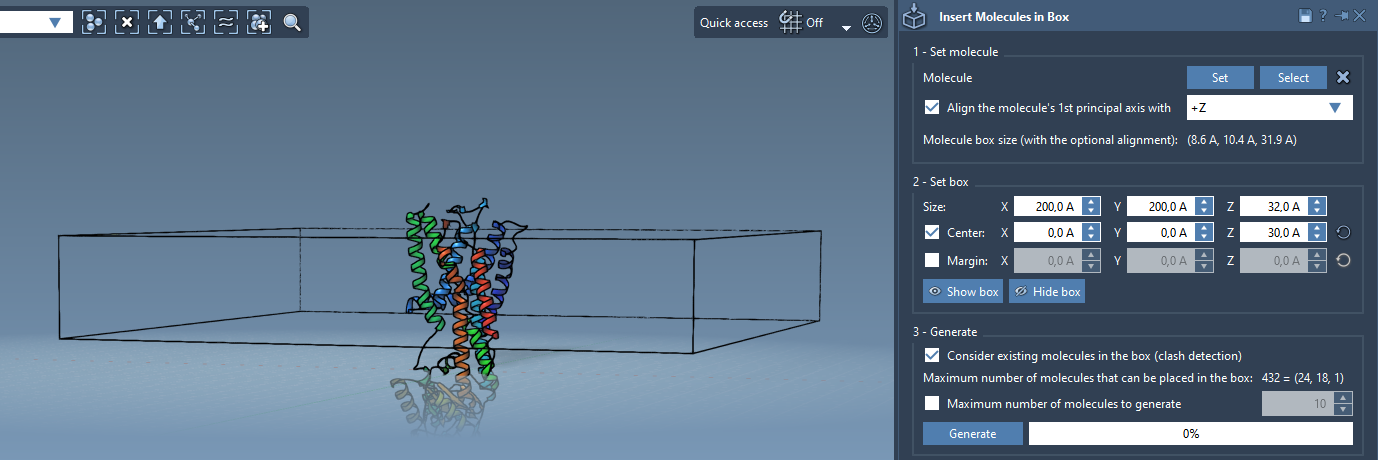
This step requires specifying the size and location of the box relative to the protein:
- Activate Center to center the box around the origin (or shift manually to center on the protein).
- Adjust the dimensions in X, Y, and Z to enclose the region where you want the lipids to be placed. For a monolayer, the Z-dimension should fit a single row of lipids.
- Optionally, set a margin between lipids to avoid unrealistic overlaps or too-dense packing.
Generating the Lipid Layer
Once the setup is complete:
- Enable Consider existing molecules in the box to avoid overlapping lipids with the protein.
- Click Generate. SAMSON will fill the available space in the box with non-overlapping copies of the lipid, respecting the box boundaries and existing molecules.
After a few seconds, you’ll see a lipid layer surrounding the protein structure.

Optional: Add a Second Monolayer
To construct a lipid bilayer:
- First add the upper layer using alignment
+Z. - Shift the box position downward along the Z-axis.
- Add a second layer with alignment
-Z.
This creates a realistic bilayer environment, enabling more representative simulations of membrane proteins.
Conclusion
With the Molecular Box Builder in SAMSON, building lipid layers around proteins becomes a visual, interactive experience—no scripts or external processing required. This workflow offers a fast track to simulation system preparation, from uploading your protein and lipid, to generating configurations ready for minimization and dynamics with tools like GROMACS.
Want to replicate this workflow or explore similar ones? Visit the full documentation here: Molecular Box Builder Documentation
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at samson-connect.net.





