Searching for the right molecule in a library can be a slow and frustrating process—especially when you’re dealing with substructures. Maybe you’re designing a compound with a specific functionality, or maybe you want to filter out only the molecules that contain both a fluorine and a nitrogen atom. Doing this manually? Time-consuming. Writing scripts? Not everyone’s favorite. The SMILES Manager in SAMSON provides a practical solution with a visual, interactive interface that simplifies advanced substructure search workflows.
This type of substructure-based filtering can be tricky, particularly when you’re screening large libraries. Fortunately, built on RDKit, the SMILES Manager helps carry out complex structural queries without code.
How It Works
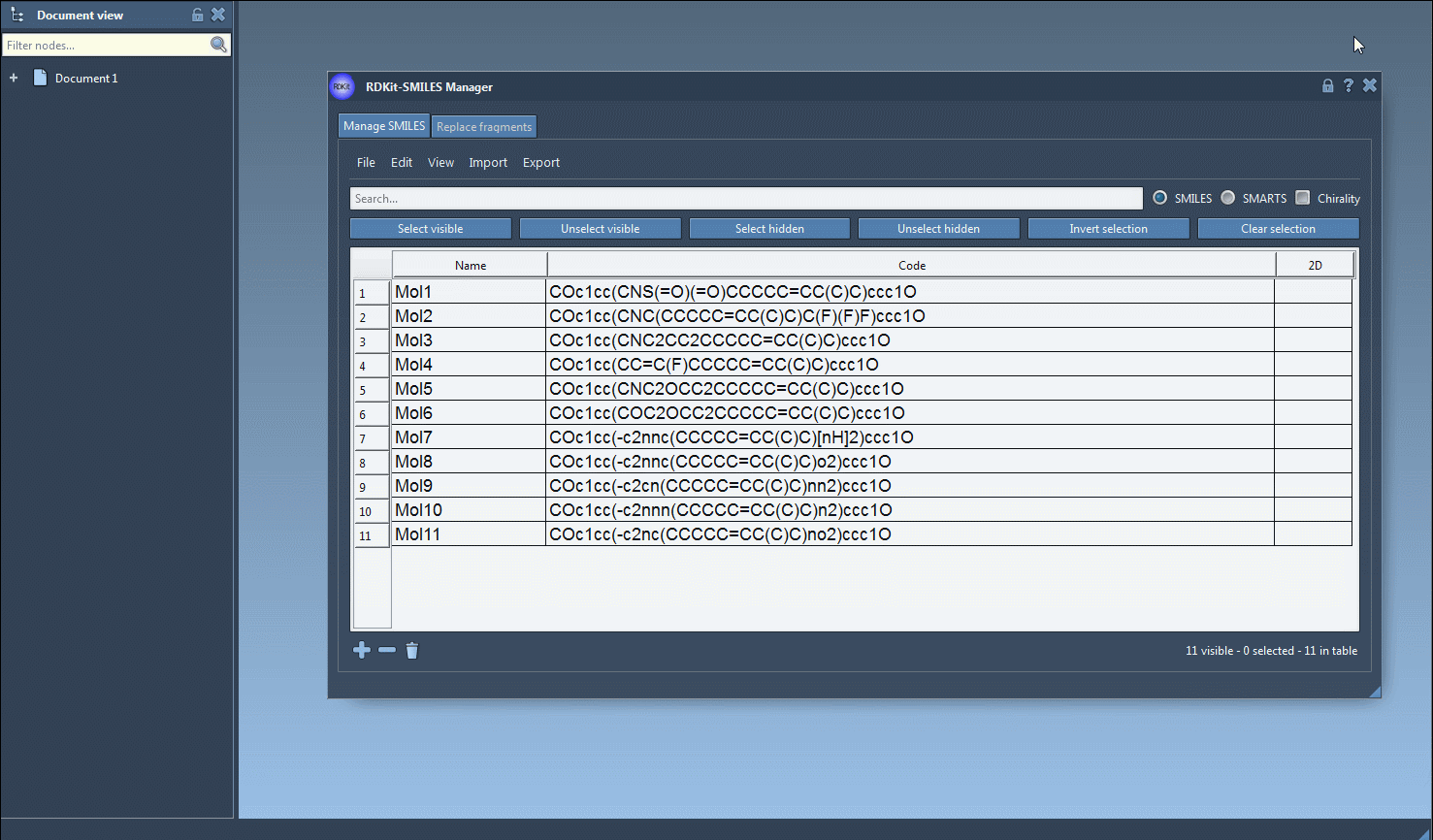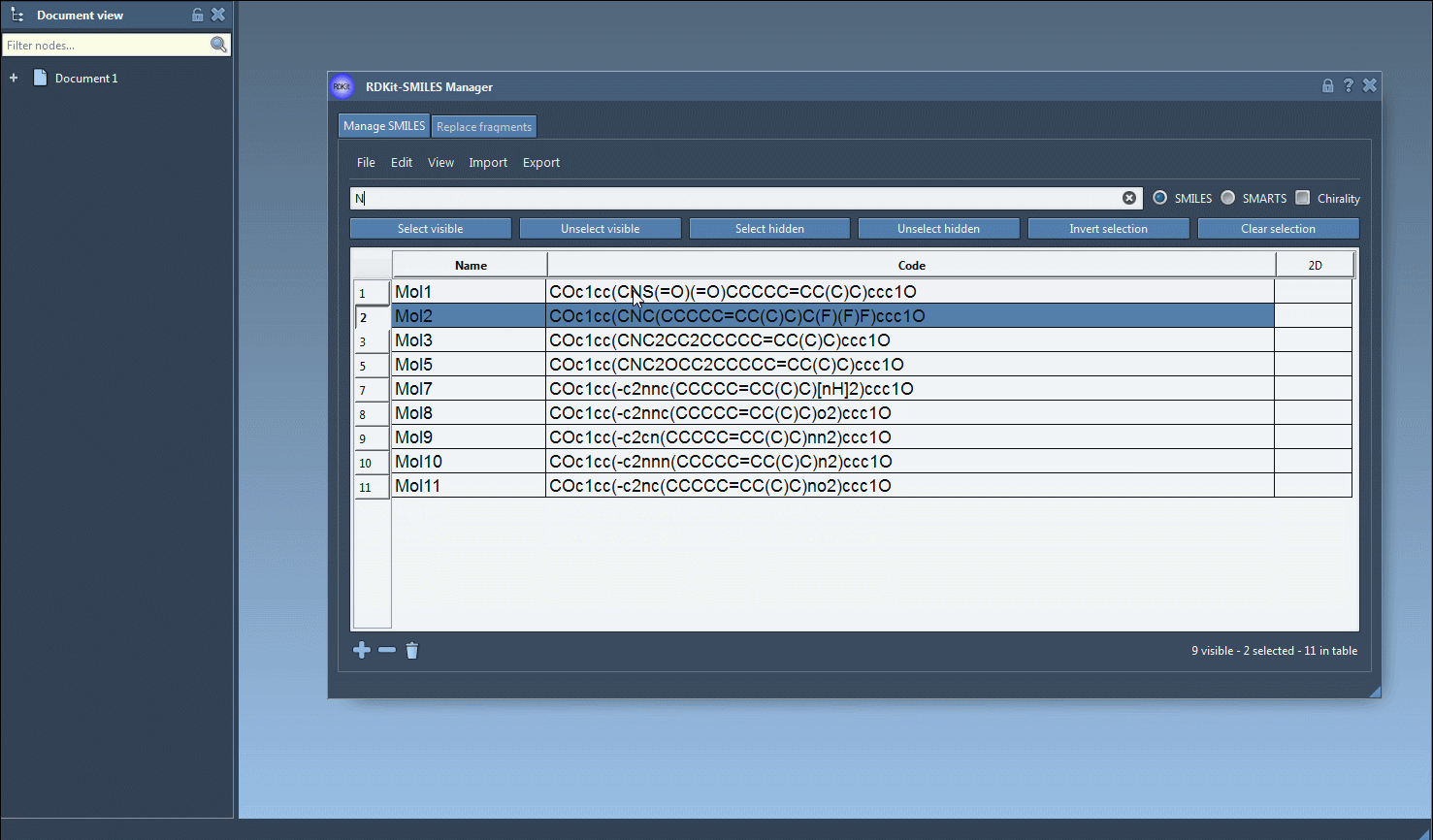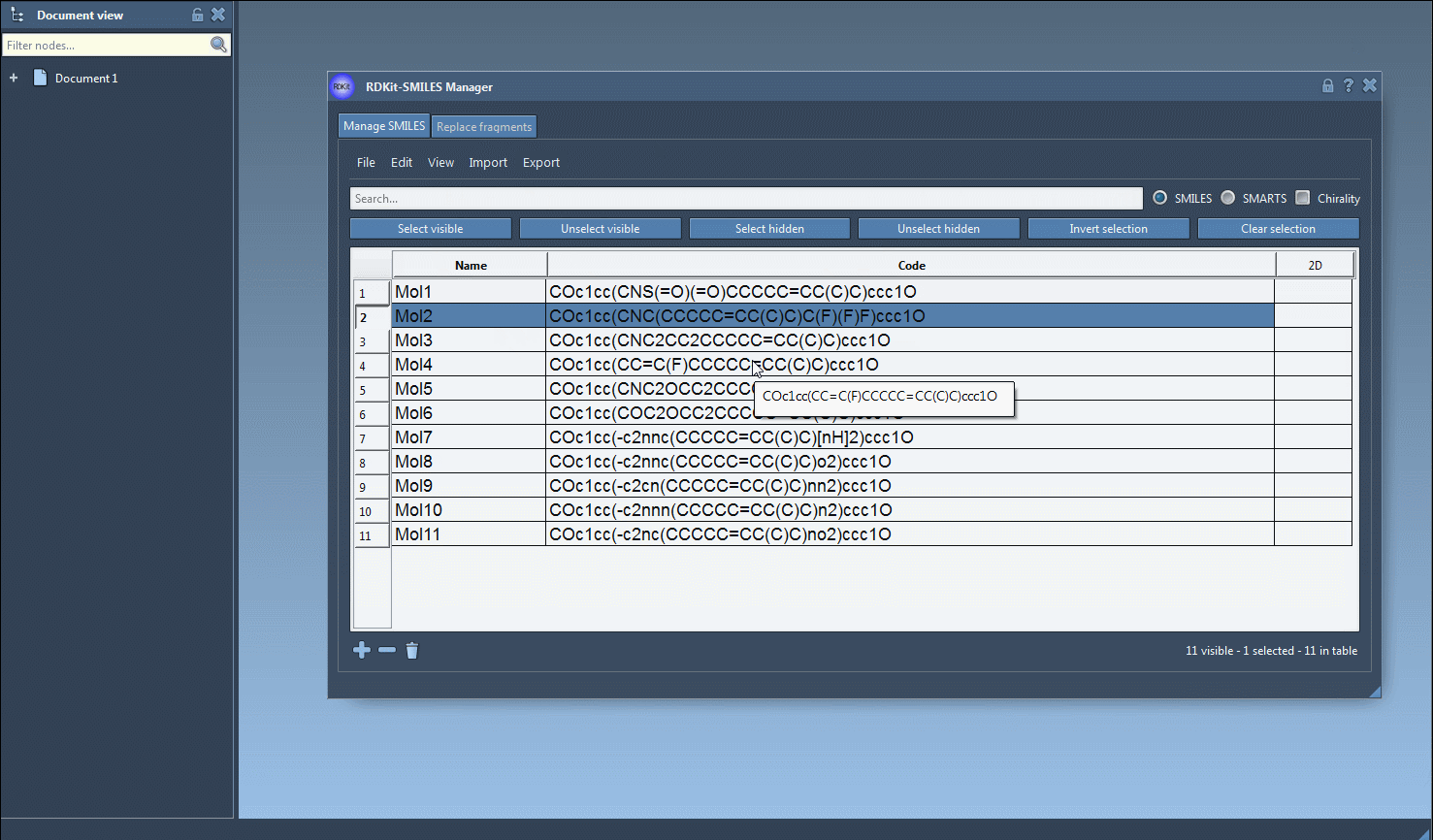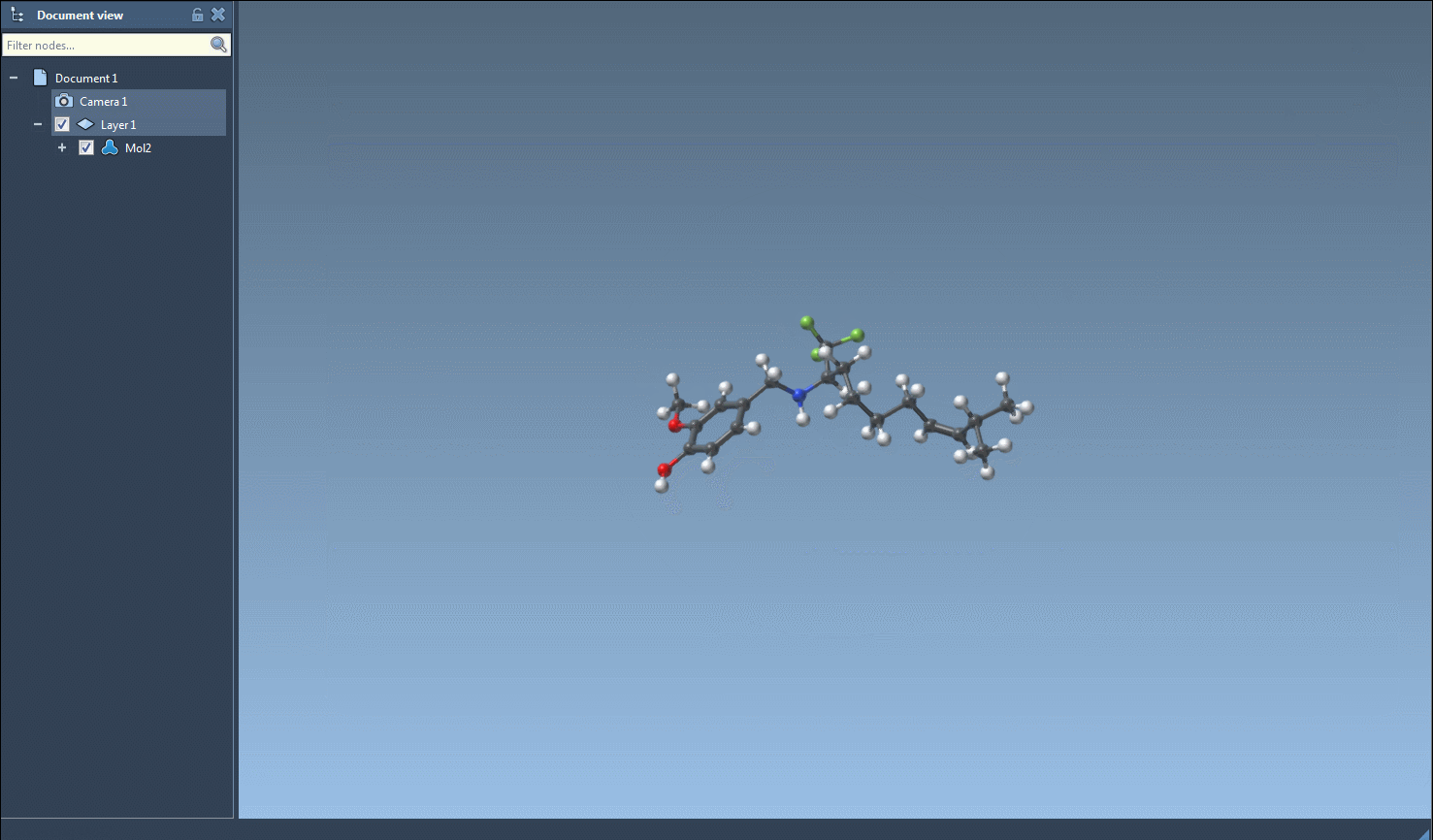
Let’s say you are only interested in those SMILES strings that contain both a fluorine atom and a nitrogen atom—but not necessarily next to each other in the molecular structure. Here’s how you can do it using a straightforward selection workflow:
- In the Search bar, enter
Fto retrieve all visible molecules containing a fluorine atom. - Click Select visible to save this set of results as a selection.
- Clear the search bar to return to the full list. The initial fluorine-containing molecules remain selected.
- Now search for
Nto list molecules that contain nitrogen atoms. - Click Unselect hidden to retain only those molecules that were both selected earlier and are visible now—in this case, molecules with both fluorine and nitrogen.
This creates a logical “AND” operation using nothing but a graphical interface—no programming required. It’s ideal for researchers reviewing large compound libraries, medicinal chemists doing scaffold hopping, or students who are still becoming familiar with structure queries.
A Closer Look
As shown in the image above, the interactive interface highlights selected molecules in real time. After narrowing down your compounds, you can easily generate 2D or 3D structures, export them, or even use them as starting points for further fragment replacements.
Note: By default, RDKit does not consider stereochemistry in substructure searches. This behavior can be changed by enabling chirality settings.
Fewer Clicks, Sharper Molecules
The beauty of this system is how accessible it is. For users unfamiliar with command-line cheminformatics tools, the SMILES Manager offers an intuitive way to carry out precise molecular filtering—and integrate it into downstream design workflows. Whether you’re doing QSAR studies or lead optimization, structure-based screening is now easier to manage.
To learn more about using the SMILES Manager for substructure search and other tasks like converting SMILES to 3D models and replacing fragments, check out the full documentation: Using the SMILES Manager.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





