When building or modifying molecular structures, one of the most frequent roadblocks in molecular modeling is the difficulty of adapting molecular topologies on the fly. Typically, breaking bonds or introducing new connections means re-running perception algorithms or manually adjusting atom types. Such interruptions not only disrupt workflow, but also increase the risk of introducing inconsistencies.
This is where the Interactive Modeling Universal Force Field (IM-UFF) in SAMSON can help. IM-UFF extends the Universal Force Field (UFF) to support real-time changes in molecular topology, such as creating and breaking bonds or changing atom types while you manipulate the system. No need to pause your simulation or restart typization—IM-UFF makes these changes continuous and responsive.
Smooth Transitions During Editing
Traditionally, molecular modelers must manually define when bonds form or break, or rely on algorithms that assume a fixed connectivity. IM-UFF changes that: it adapts bond orders and atom types based on real-time atom positions. If you move an atom slightly, the system adjusts while preserving current bonds. If you move the atom far enough away from its bonded neighbors, bonds are broken and the molecular topology updates automatically. Similarly, bringing atoms close together may result in bond formation. This process supports intuitive molecular editing and prototyping, guided by physically-based interatomic forces.
How It Works in Practice
Here’s how to try it in SAMSON:
- Create or open a molecular system in SAMSON.
- Add a simulator via Edit > Simulate > Add simulator (or use
Ctrl+Shift+Mon Windows /Cmd+Shift+Mon macOS). - Select Interactive Modeling Universal Force Field.
- Choose a state updater (for example, the FIRE relaxer).
- Start the simulation and move atoms with the mouse.
When the simulation is running, watch the bond network adjust as you displace atoms. The total energy and its components are visible in the simulation window so you can understand what is driving changes in structure. You can even add or delete atoms on the fly—IM-UFF will manage the perception of new bond orders automatically.
Key Options That Influence Behavior
Make sure the following two options are unchecked to leverage full interactivity:
- Static topology (UFF only) – Unchecking this enables dynamic topology changes.
- Keep vdW for manipulated – Disabling this option improves usability when you’re manually moving atoms, as it ignores van der Waals repulsion for the atom you’re actively manipulating.
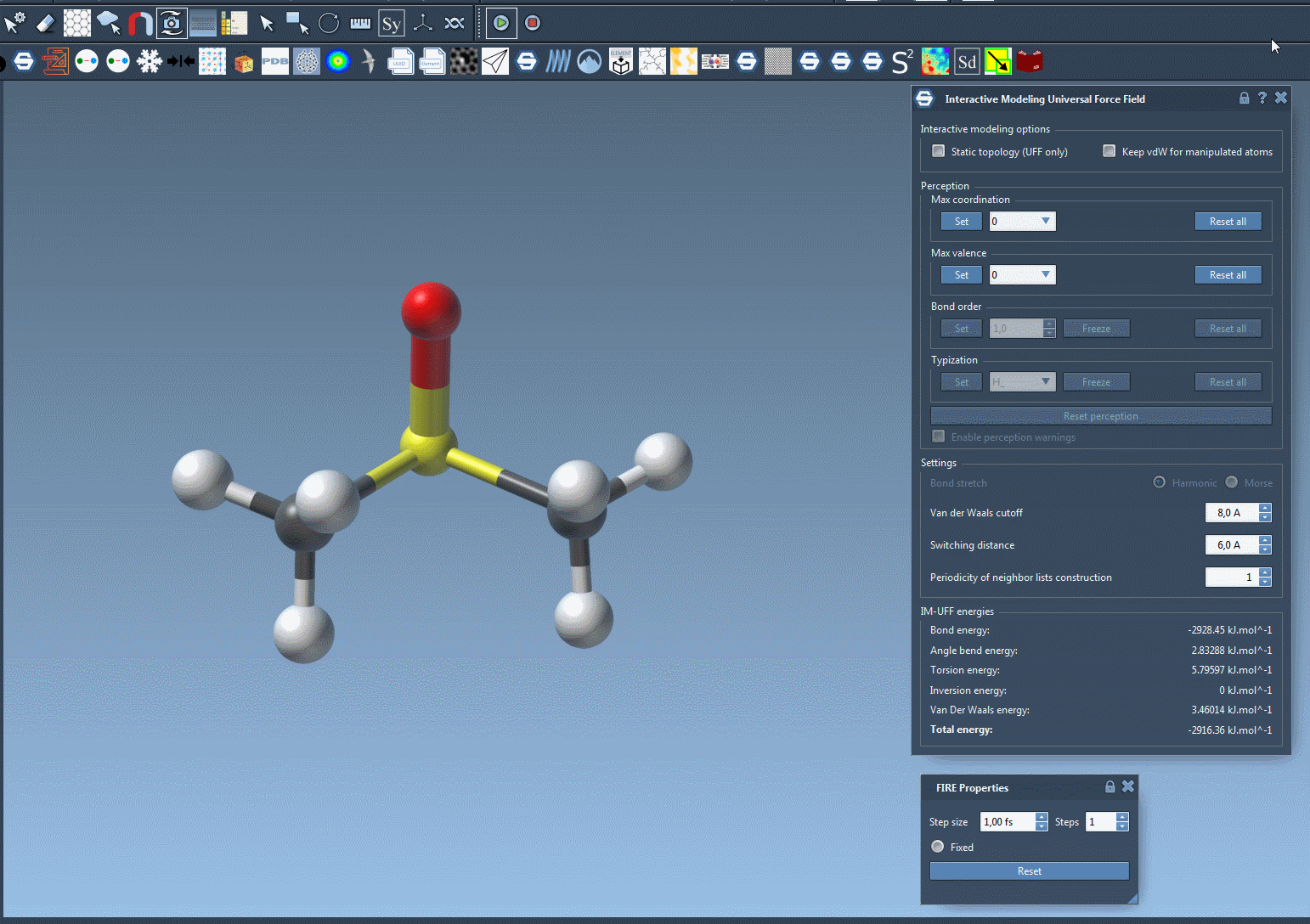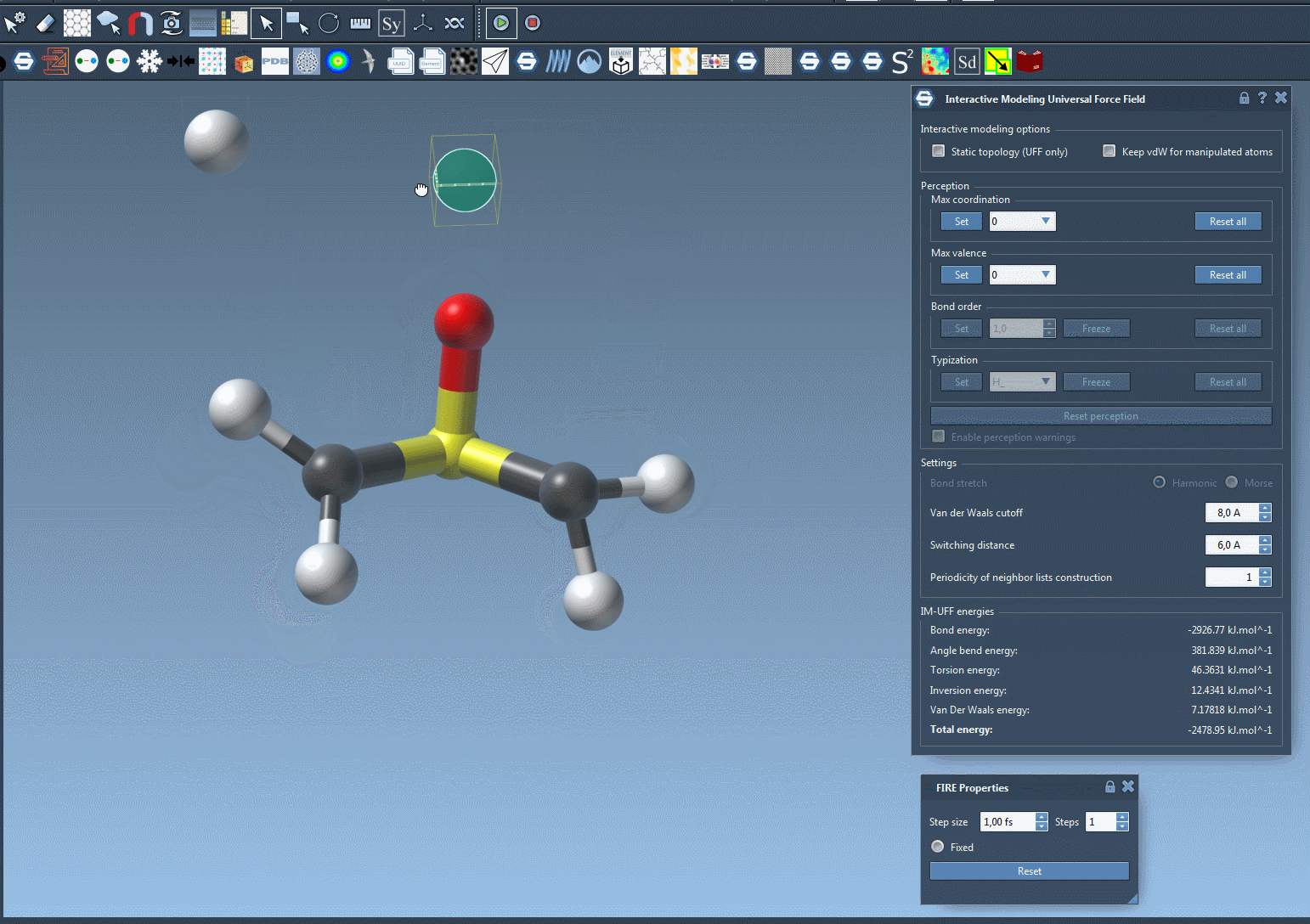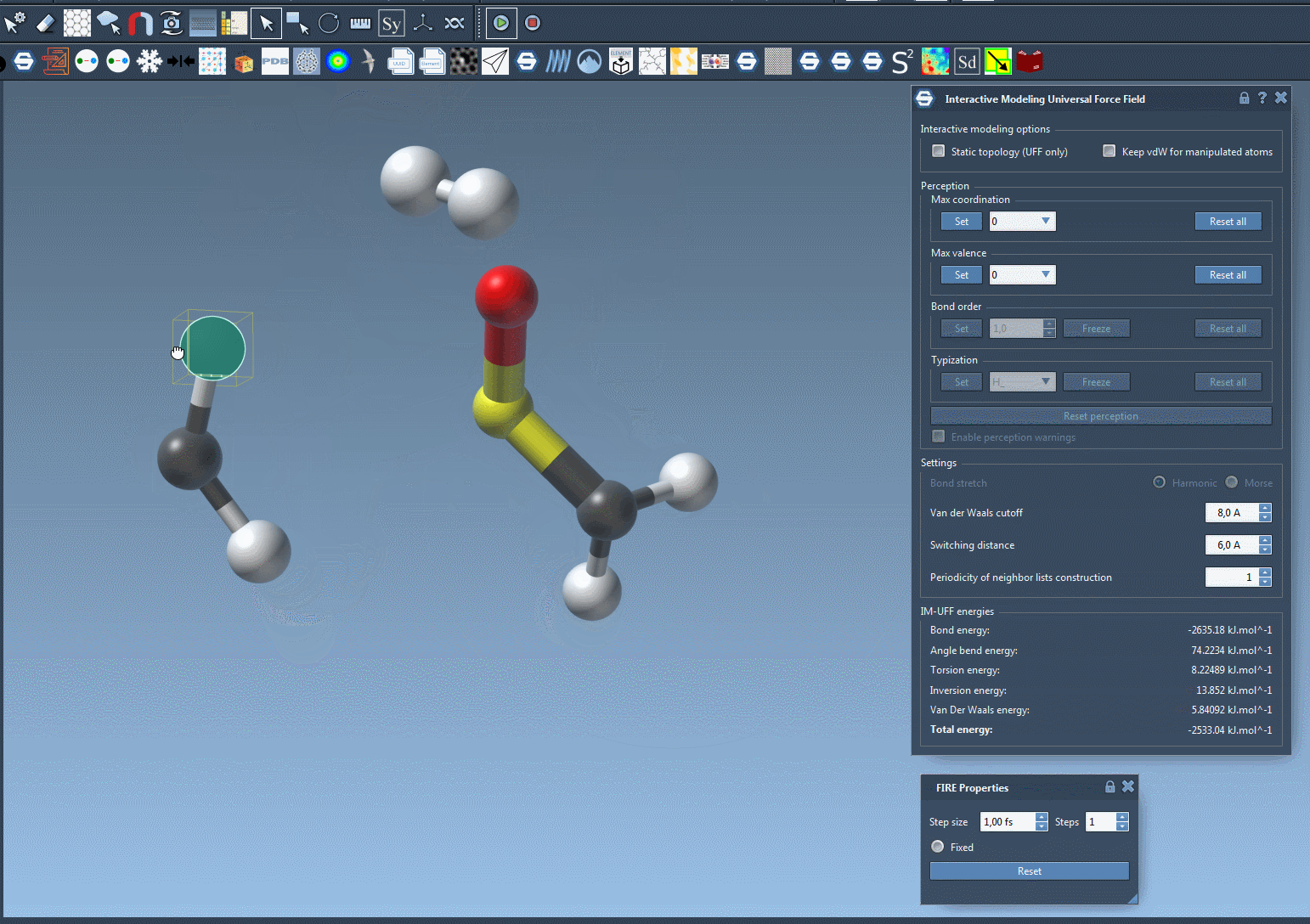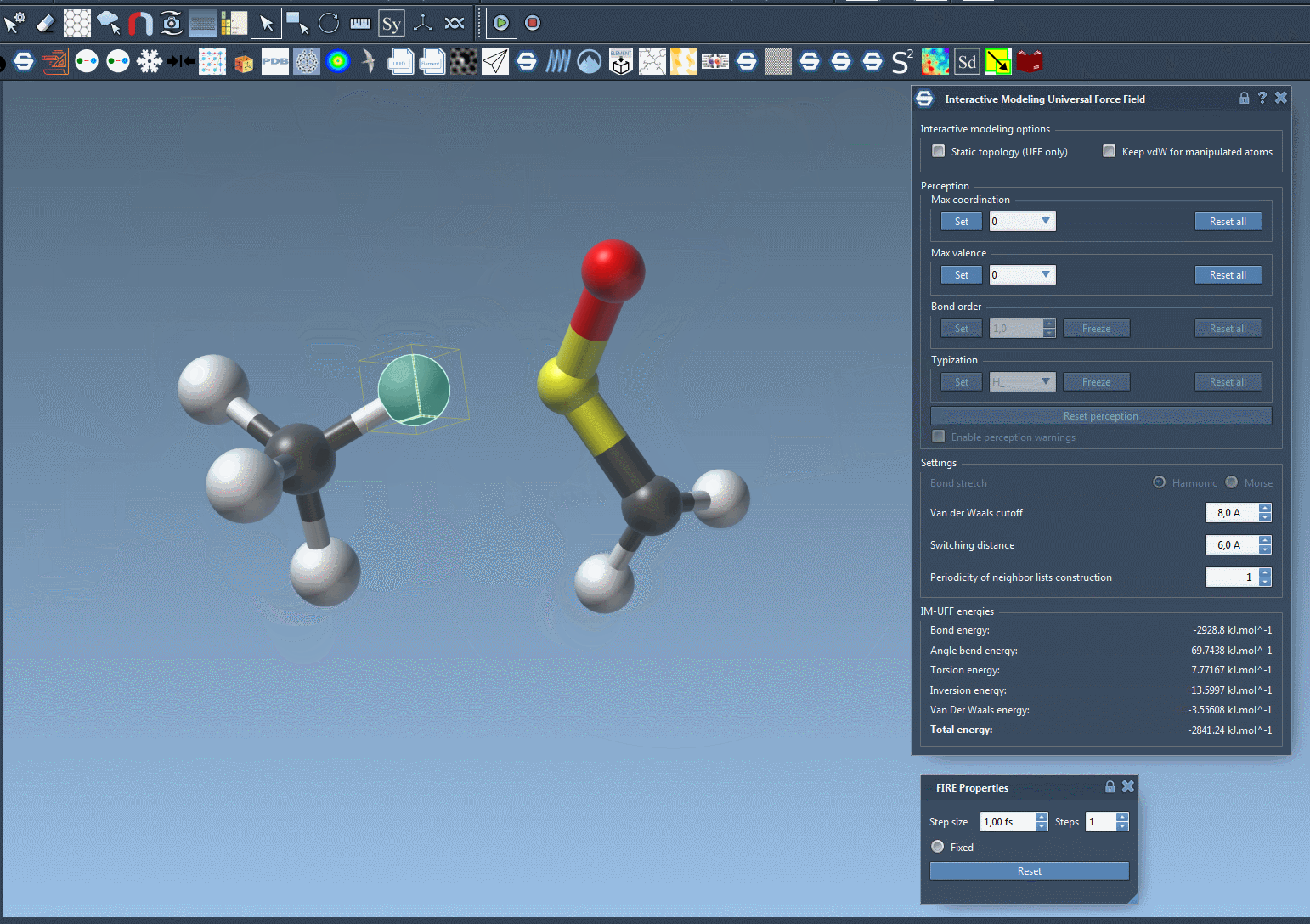
Why This Matters
For chemists, students, engineers, and anyone designing molecular systems, the ability to freely edit molecular structures without worrying about bond definitions is a significant time-saver. It lowers the barrier to experimentation and supports a more natural modeling workflow.
To learn more, see the full documentation page here: https://documentation.samson-connect.net/tutorials/uff/im-uff/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





