Searching for specific substructures across a library of molecules is a routine task in cheminformatics, but it can be tedious and error-prone when working with large datasets. If you’ve ever found yourself combing through countless SMILES strings looking for specific functional groups or atomic patterns, SAMSON’s SMILES Manager may help streamline your workflow.
This blog post walks you through an overlooked but practical feature: the improved substructure search available in the SMILES Manager Extension of SAMSON. It lets you combine multiple search criteria to accurately filter molecules that contain specific patterns—without needing scripts or third-party tools.
Why Substructure Search Matters
Substructure matching allows you to identify molecules that contain a certain functional group or atomic arrangement. It’s especially useful when designing or narrowing down a screening set, analyzing SAR trends, or simply validating a chemical database. What’s better: no coding required.
How It Works in SAMSON
The updated substructure search in the SMILES Manager works through step-wise filtering directly within the interface.
Here’s an illustrated example:
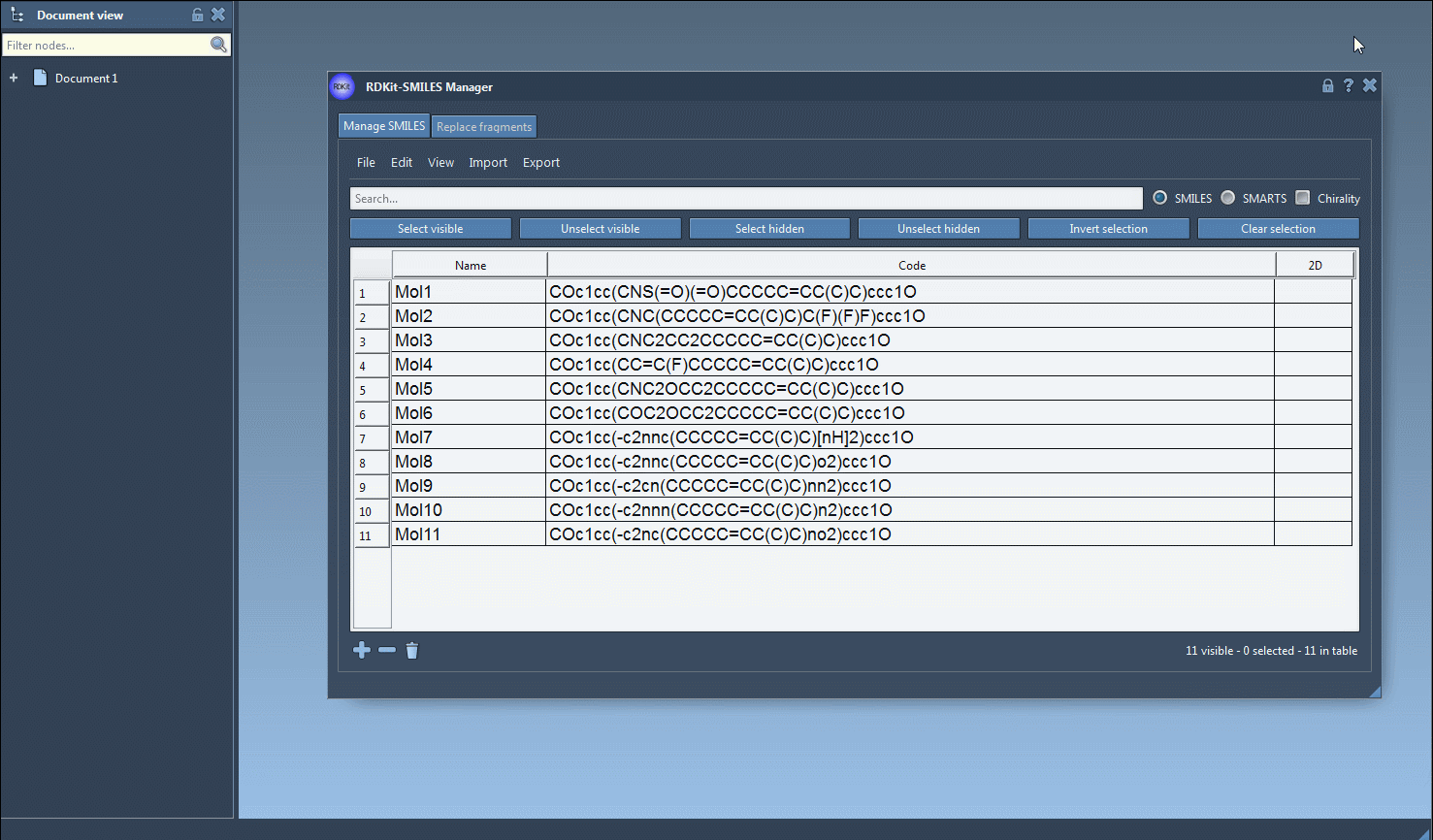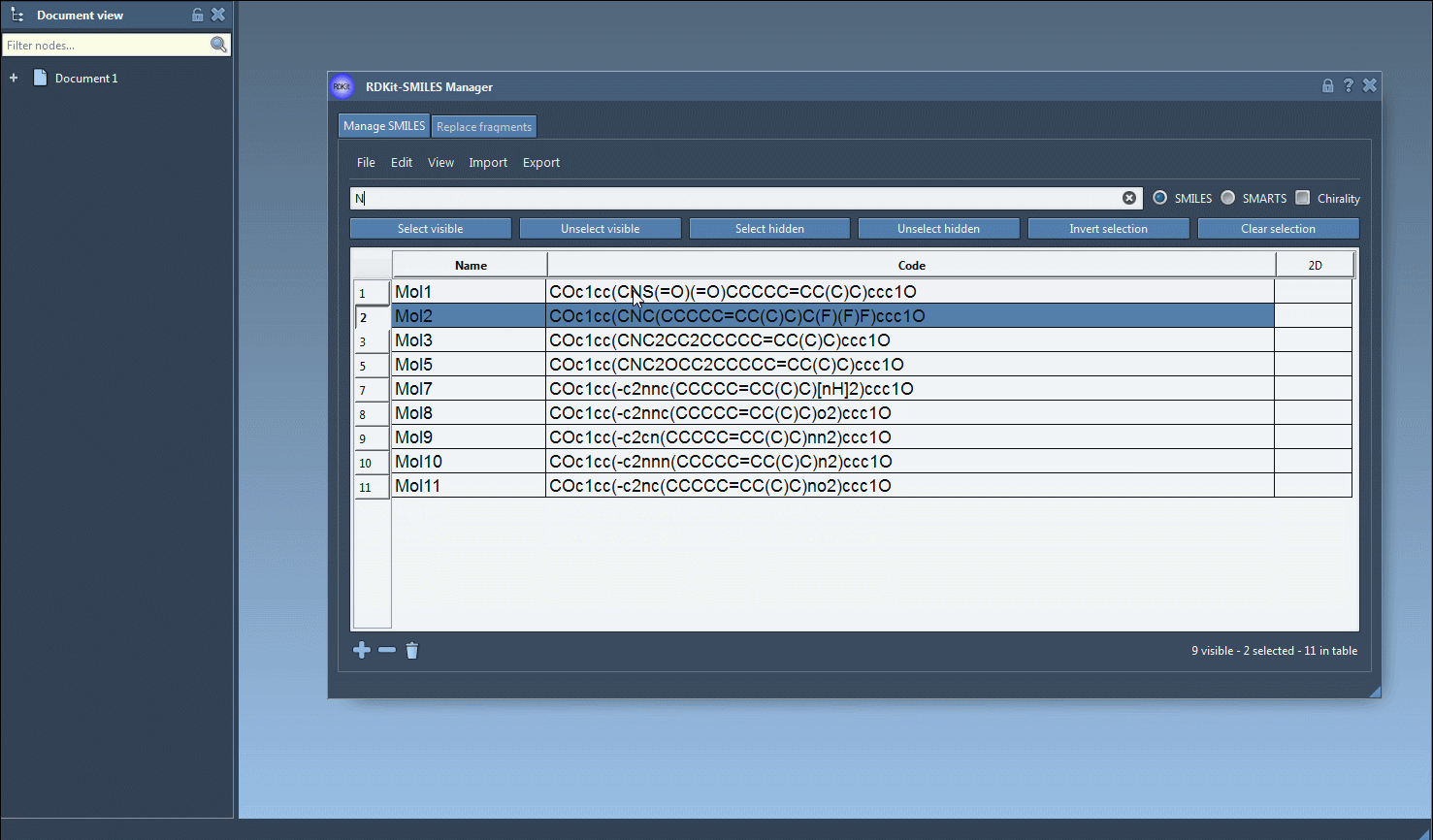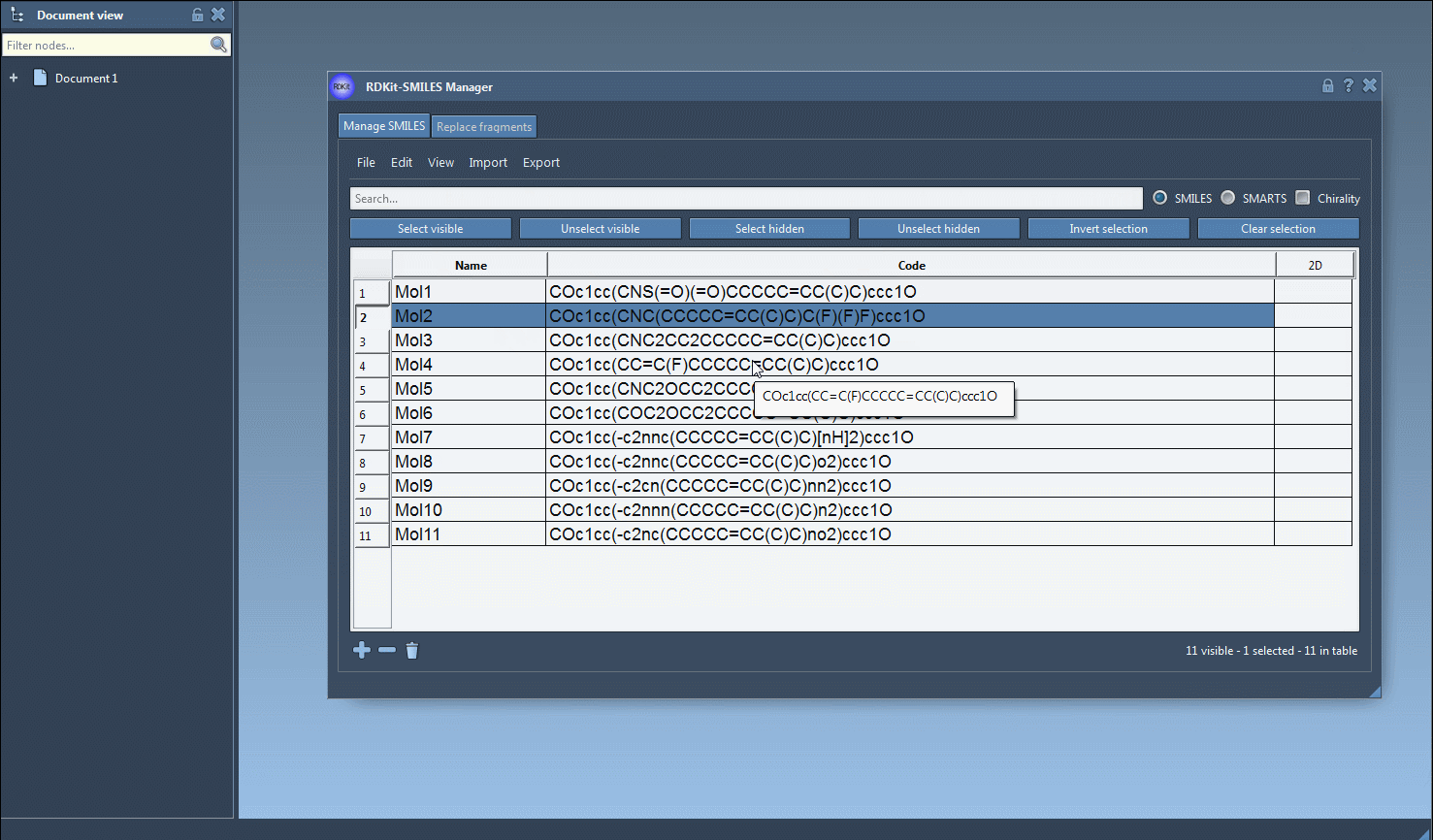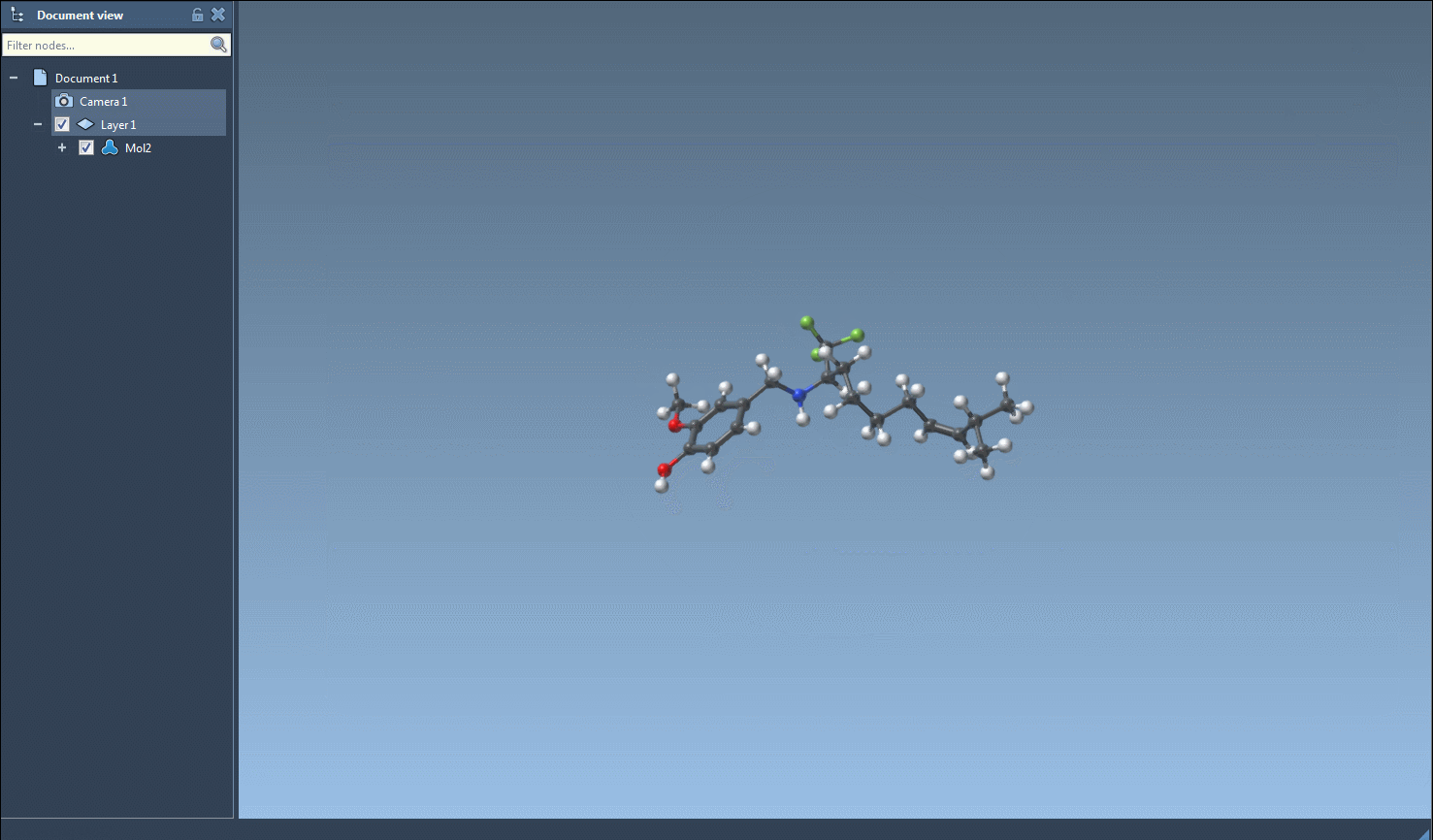
- Step 1: You load a list of molecules using their SMILES representations.
- Step 2: Enter a substructure pattern; for instance,
Fto filter for molecules with a fluorine atom. Click Select visible. These are now selected. - Step 3: Clear the search, type a new pattern—say,
Nfor nitrogen—and observe which of the previously selected molecules remain visible. - Step 4: Click Unselect hidden to keep only those SMILES that match both patterns.
This method allows you to iteratively refine your selection using logical intersections—without typing any Boolean logic.
Where This Helps
Compared to basic text search, this approach offers several benefits:
- Efficiency: No need to manually inspect or script the filtering logic.
- Visualization: Matched molecules are visible with 2D depictions, making it easy to confirm hits visually.
- Refinement: You can continue building your set by applying more criteria step by step.
Note: This feature uses RDKit’s substructure matching by default, and chirality is not considered unless explicitly enabled via settings. So you can work quickly unless stereochemistry is important for your analysis.
Next Steps
Once your selection is complete, you can:
- Generate 3D structures of the filtered molecules.
- Export them into SAMSON Documents or image grids for reporting.
- Save SMILES lists or SVG depictions.
If you’ve been relying on lengthy command-line scripts or external tools for substructure filtering, this SAMSON feature might simplify many of your exploratory tasks.
To learn more or explore other SMILES Manager capabilities such as fragment replacement or 3D generation, visit the official documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at samson-connect.net.





