One of the recurring challenges for molecular modelers working with SMILES files is identifying and isolating molecules that contain specific substructures. Searching by substructure matters—a lot—when you’re building small molecule libraries, designing focused chemical series, or preparing datasets for further screening.
But let’s be honest: many molecular editors make this process tedious. Searching means reloading files, manually building subsets, or—worse—losing the previous selection as soon as you run another filter.
If this sounds familiar, you may find the SMILES Manager Extension in SAMSON particularly helpful. Thanks to a clever combination of selection and visibility tools, it lets you chain searches without breaking your workflow. Here’s how it works.
Progressive Substructure Search in SMILES Manager
Let’s say you want to pick out only the molecules that contain both a fluorine F and a nitrogen N atom, using exact substructure patterns. Here’s how to do it with confidence.
- Search for your first substructure (e.g.,
F). Enter it in the search bar and press Select visible. This grabs all molecules currently shown that contain a fluorine atom. - Clear the search field. Your fluorine-bearing molecules remain selected—even though you’re no longer filtering.
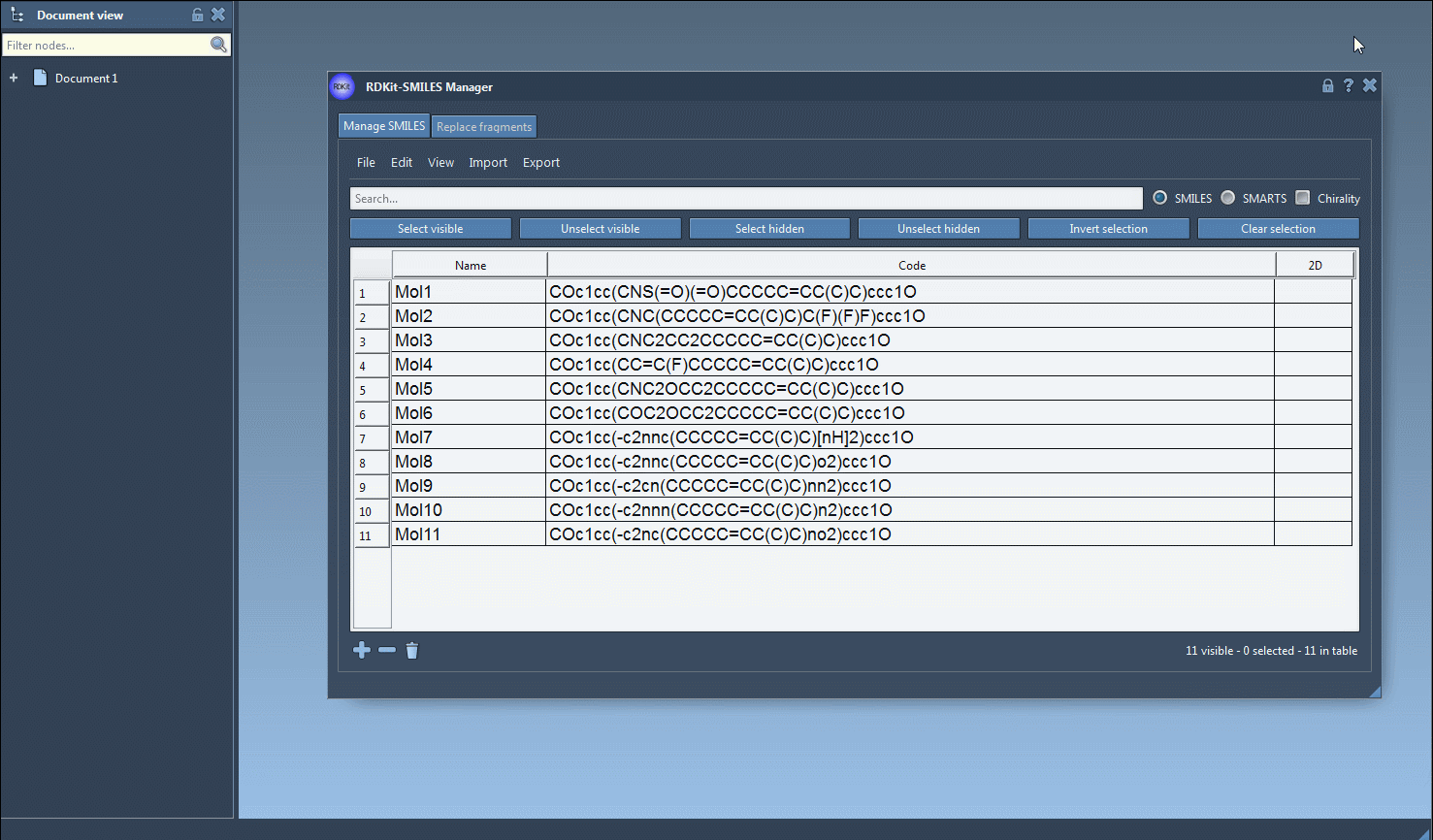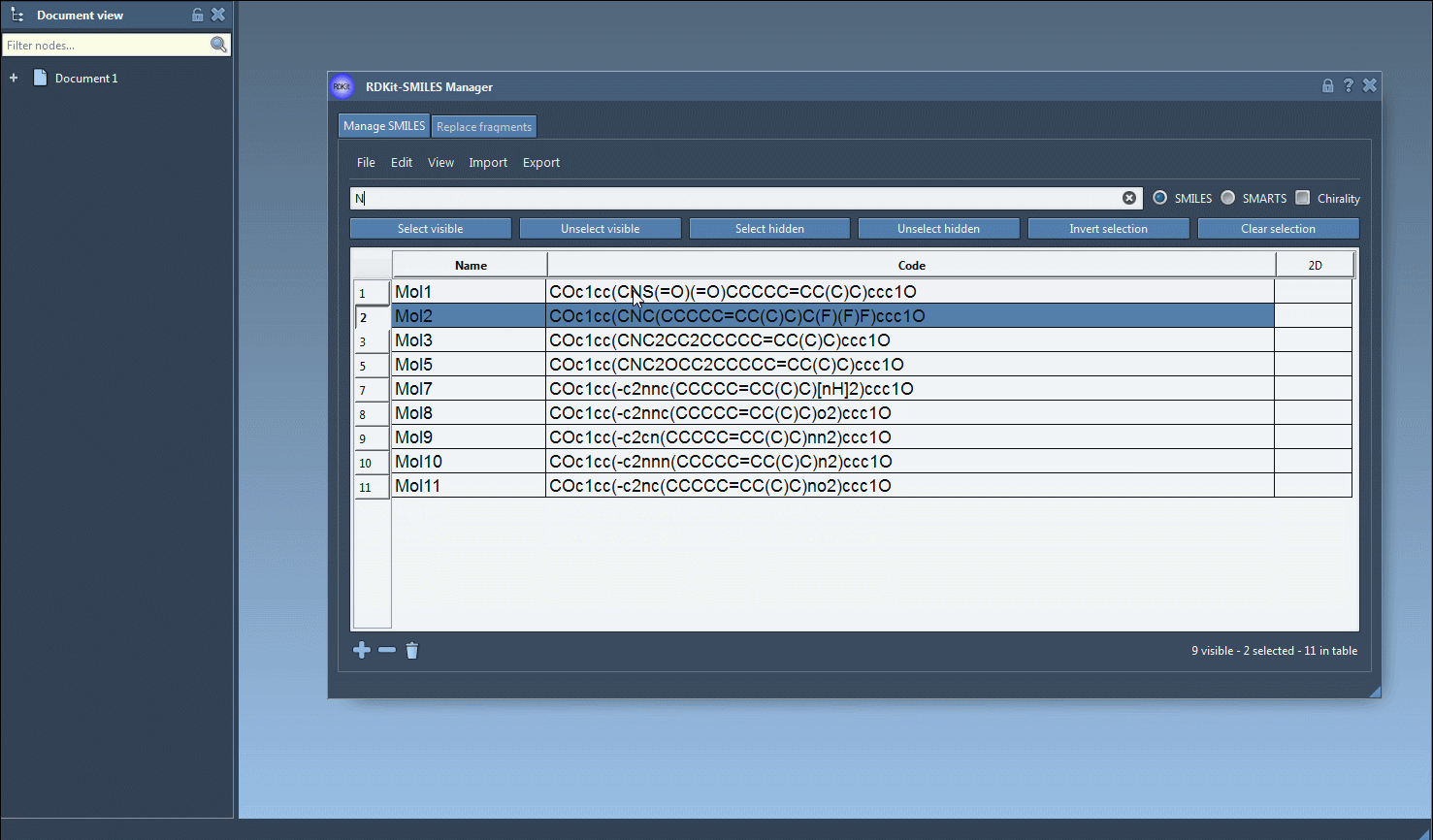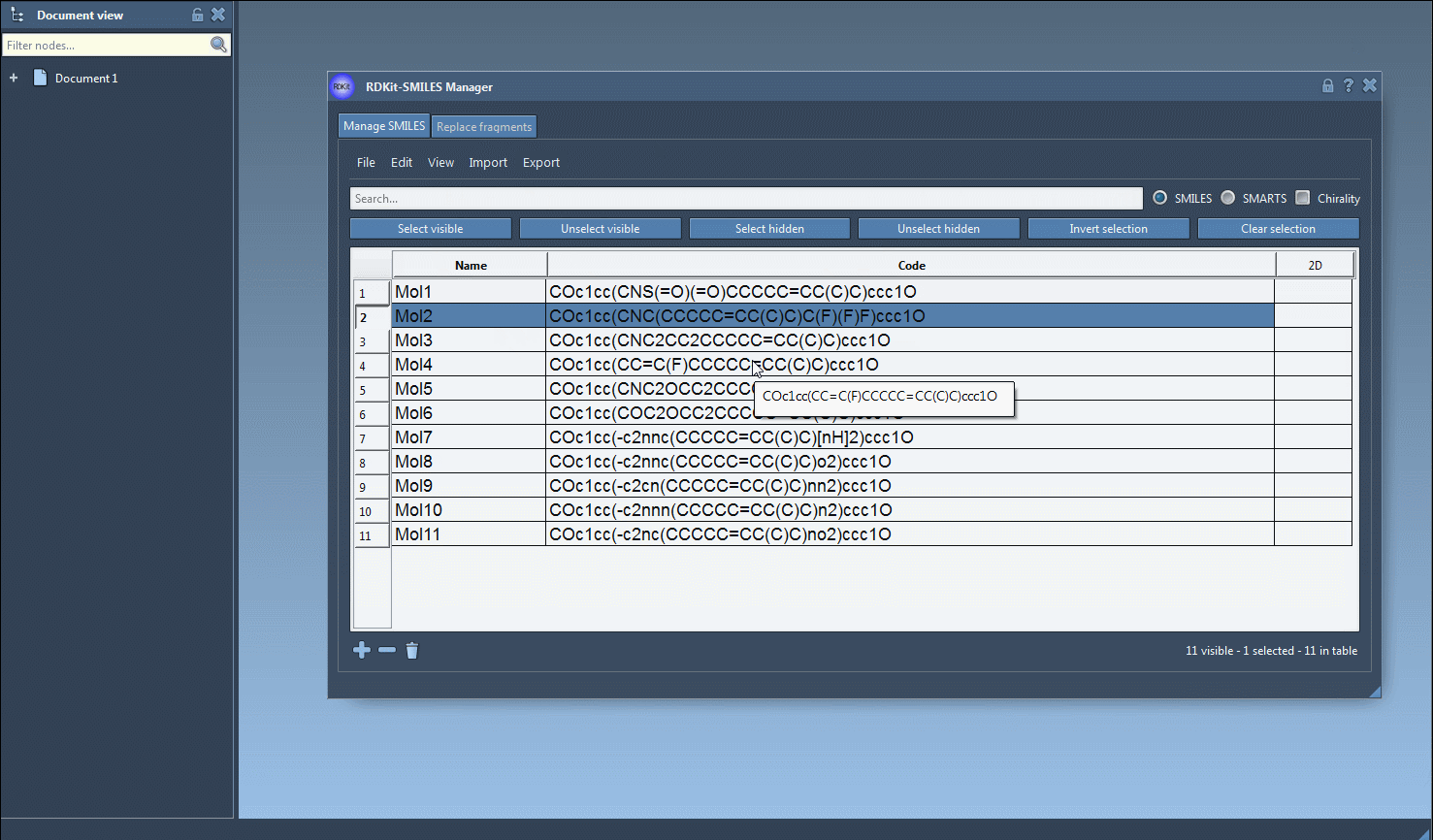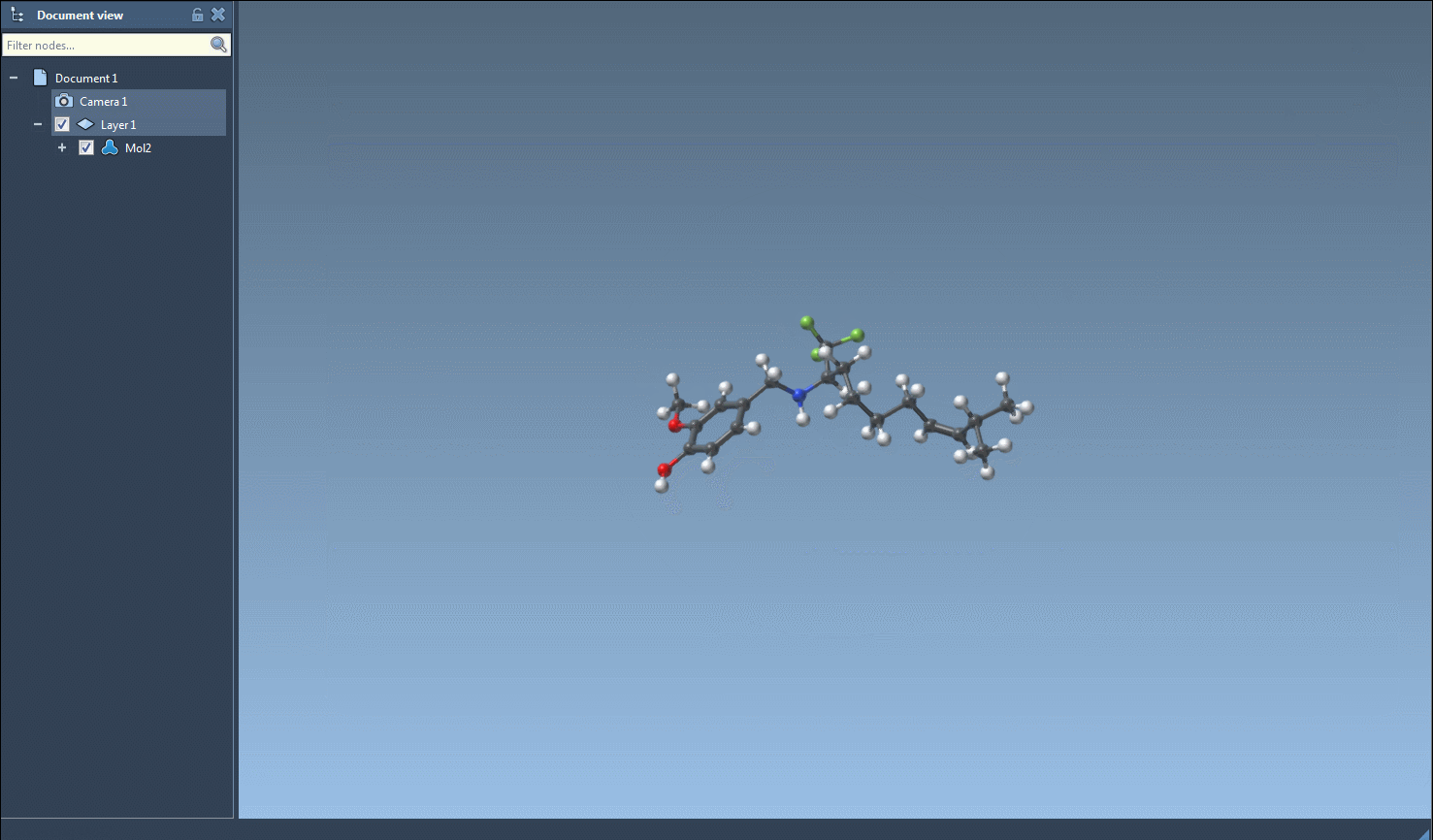
- Now search for your second substructure (e.g.,
N), and observe which of your already selected molecules are still visible. This filters the selected set by those that also contain nitrogen. - Click on Unselect hidden. You’re now left with only the molecules that include both fluorine and nitrogen.
This method allows for compound filtering without losing the original selection. It’s simple, fast, and much less frustrating than rebuilding selections manually or re-running structural queries in external tools.
Additional Notes
By default, RDKit (on which SMILES Manager is built) performs substructure matching without taking stereochemistry into account. If you’re working with chiral centers and need to preserve stereochemical information, you’ll want to enable chirality-aware matching (this is possible in RDKit, although the documentation does not show the full details here).
This feature is especially useful when triaging large combinatorial libraries or performing focused searches during structure expansion. It gives you a fast, visual, and interactive way to refine your hit lists, whether you’re working with a handful of molecules or thousands.
To learn more about managing and searching SMILES strings in SAMSON, check out the full documentation page here: https://documentation.samson-connect.net/tutorials/smiles-manager/using-the-rdkit-smiles-manager/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





