Tracking how ligands move through molecular systems — such as during unbinding or along diffusion pathways — is often a challenging task for molecular modelers. The movement can be complex, and visualizing it clearly across multiple frames of a trajectory is not always straightforward.
This is where the Pathlines visual model in SAMSON comes in. It lets you display the center-of-mass (COM) trajectory of selected atoms along a predefined path. This approach is ideal for highlighting ligand dynamics, especially in systems with extensive molecular motion such as conformational changes in proteins.
Why Use Center-of-Mass Pathlines?
- They reduce visual clutter by representing the average motion of atom groups rather than individual atoms.
- They help identify unbinding paths, diffusion trends, and domain shifts in biomolecular simulations.
- They make it easier to communicate complex motions in presentations or publications.
Step-by-Step: Visualizing Ligand Unbinding
Let’s take a look at an example — visualizing the unbinding of the ligand Thiodigalactosid (TDG) from the protein Lactose permease (PDB: 1PV7) using a sample project and the Pathlines extension in SAMSON.
1. Load the Sample System
You can start by downloading the preconfigured system that includes the protein, ligand, and pre-computed paths. Simply paste the following document link when prompted in SAMSON:
https://www.samson-connect.net/documents/046f1acd-c799-40f6-8185-cb4847eff795

2. Select the Ligand and Paths
In the Document view of SAMSON, select the ligand (TDG) from the sample and the path(s) you want to visualize. If you don’t select a specific atom group or path, SAMSON uses the entire system and all paths by default.

3. Create a Pathline Visual Model
Once your atoms and paths are selected:
- Go to
Visualization > Visual model > More... - Choose Pathline of the center of mass
- Click OK
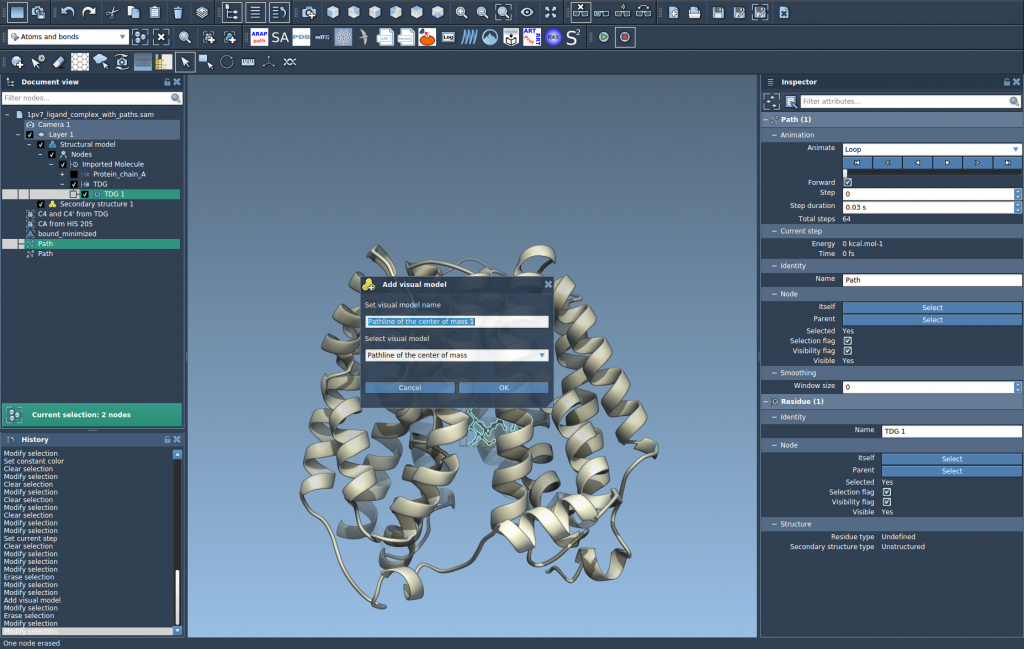
That’s it — you’ll now see a continuous line representing the COM of the ligand segment as it moves along the selected path.
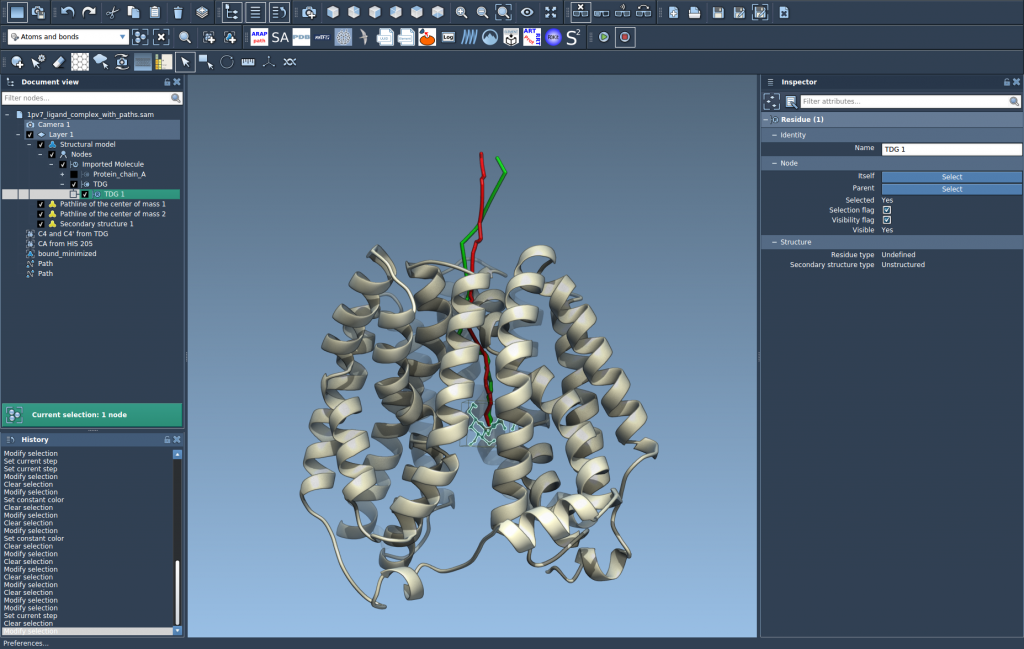
4. Customize and Explore
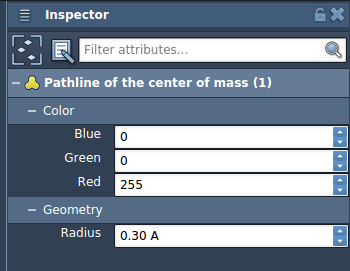
You can explore the path by double-clicking it in the Document view or by accessing its settings through right-click options. In the Inspector, you can adjust attributes such as color and thickness to make the pathline clearer for communication or publication purposes.
When to Use This Technique
Center-of-mass trajectory visualization is particularly useful in three cases:
- Showing ligand unbinding/rebinding routes from molecular dynamics simulations
- Analyzing large movement patterns, such as domain rearrangement in protein complexes
- Understanding trajectory trends in reaction coordinate workflows
This approach simplifies interpretation and enhances how you present your modeling results to your team or collaborators.
To learn more about setting up pathlines and tailoring them to your workflow, visit the official documentation: Pathlines documentation in SAMSON.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





