For molecular modelers working on protein-ligand interactions, refining transition pathways between binding and unbinding states is essential—but often challenging. Whether you’re studying conformational changes, binding affinity, or energy landscapes, having a more precise transition path between low-energy states directly improves the interpretability of your simulations.
One common pain point is that molecular simulations often produce paths that are noisy or physically unrealistic due to insufficient sampling or large system complexity. This is where the Parallel Nudged Elastic Band (P-NEB) method in SAMSON offers a practical and accessible refinement tool.
What is P-NEB?
The P-NEB (Parallel Nudged Elastic Band) app in SAMSON implements the climbing image nudged elastic band algorithm, which refines a pathway by minimizing energy while maintaining spacing between conformations. It’s especially useful when you already have two local energy minima and want to interpolate the transition between them more accurately.
The process can be applied both to a full path (i.e. a trajectory) and to a group of conformations (snapshots), but working with paths is often more efficient and recommended.
Start from a Path or Create One
If you begin with a set of relaxed conformations—say from a linear interpolation or tools like Ligand Path Finder—you can combine them into a path directly in SAMSON:
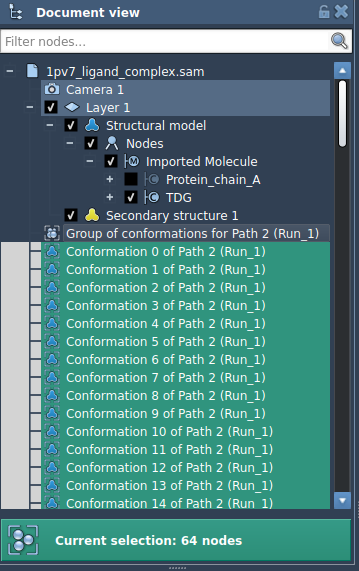
- Select the conformations in the Document view
- Right-click and choose
Conformation > Create path from conformations
Launching the P-NEB App
Once you have a path or a set of conformations ready:
- Go to Home > Apps > All > P-NEB
- Select your path or conformations in the document
- Click Run in the P-NEB interface
Before launching, configure the settings. For example:
- Spring constant: 1.00
- Number of loops: 100
- Interaction model: Universal Force Field
- Optimizer: FIRE (Fast Inertial Relaxation Engine)
- Climbing image method: leave unchecked initially
- Parallel execution: check this for better performance
- Suffix name: NEB
What to Expect
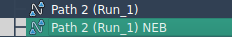
After clicking Run, SAMSON will ask whether to use existing bonds. Choose “Yes”. Then, the optimization will begin and progress updates will be shown in the status bar.

Once completed, a new path (or new conformations) will appear in the Document view, with the suffix NEB added to its name. You can visualize the refined transition using animations and inspect the energy profiles at each step.
When to Use P-NEB
If you’re modeling transitions such as ligand unbinding, loop movement, or side-chain flipping, and you already have either a coarse path or sampled conformations, P-NEB can significantly improve realism. It sharpens noisy segments and helps identify energy barriers along the path.
To learn more and follow a complete tutorial, visit the official SAMSON documentation: P-NEB Documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON here.





